
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
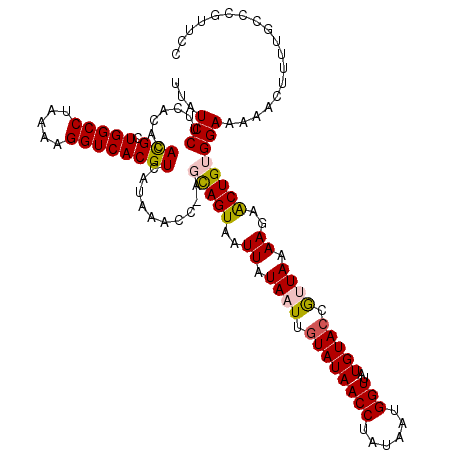
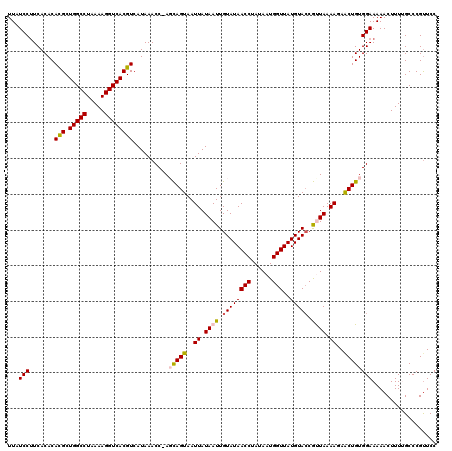
| Location | 8,500,439 – 8,500,593 |
| Length | 154 |
| Max. P | 0.973851 |

| Location | 8,500,439 – 8,500,559 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.89 |
| Mean single sequence MFE | -24.04 |
| Consensus MFE | -21.30 |
| Energy contribution | -21.42 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.889790 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8500439 120 + 22407834 UUAUCCUUCACACACGCUGGCCUAAAAGGUCACGUCAUAAACCAAGCAGUAAUUAUAAUUGUAUAACCUAUAAUGGUUAUGUAGCGUUAAAAGAACUGUGGAAAAACUUUUGCCCGCUCC ...(((.......(((.(((((.....))))))))..........(((((..((.(((((((((((((......)))))))))..)))).))..)))))))).................. ( -24.30) >DroSec_CAF1 9876 119 + 1 UUAUCCUUCACACAUGCUGGCCUAAAAGGUCACGUCAUAAACC-AGCAGUAAUUAUAAUUGUAUAACCUAUAAUGGUUAUGUACCGCUAAAAUAACUGUGGAAAAACUUUUGCCCGUUCC ......((((((.(((((((((.....))))).).))).....-(((.(((..........(((((((......)))))))))).)))........)))))).................. ( -21.80) >DroSim_CAF1 9085 119 + 1 UUAUCCUUCACACACGCUGGCCUAAAAGGUCACGUCAUAAACC-AGCAGUAAUUAUAAUUGUAUAACCUAUAAUGGUUAUGUACCGCUAAAAGAACUGUGGAAAAACUUUUGCCCGUUCC .............(((.(((((.....))))))))........-(((.(((..........(((((((......)))))))))).)))....((((.(..(((....)))..)..)))). ( -23.30) >DroEre_CAF1 9495 119 + 1 UUAUCCUUCACACACGCUGGCCUAAAAGGUCACGUCAUAAACC-ACUAGUAAUUAUAAUUGUAUAACCUAUAAUGGUUAUGUACCAUUAAAAGAGCUGUGGAAAAACUUUAGCCCGUUCC .............(((.(((((.....))))))))......((-((.(((..((.((((.((((((((......)))..))))).)))).))..)))))))................... ( -24.60) >DroYak_CAF1 10075 119 + 1 UUAUCCAUCACACACGCUGGCCUAAAAGGUCACGUCAUAAACC-ACUAGUAAUUAUAAUUGUAUAACCUAUAAUGGUUAUGUACCAUUAAAAGAGCUGUGGAAAAACUUUGGCCCGUUCC ....(((......(((.(((((.....))))))))......((-((.(((..((.((((.((((((((......)))..))))).)))).))..)))))))........)))........ ( -26.20) >consensus UUAUCCUUCACACACGCUGGCCUAAAAGGUCACGUCAUAAACC_AGCAGUAAUUAUAAUUGUAUAACCUAUAAUGGUUAUGUACCGUUAAAAGAACUGUGGAAAAACUUUUGCCCGUUCC ...(((.......(((.(((((.....))))))))..........(((((..((.((((.((((((((......)))..))))).)))).))..)))))))).................. (-21.30 = -21.42 + 0.12)

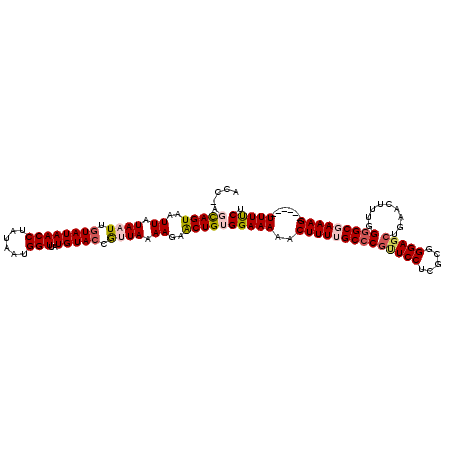
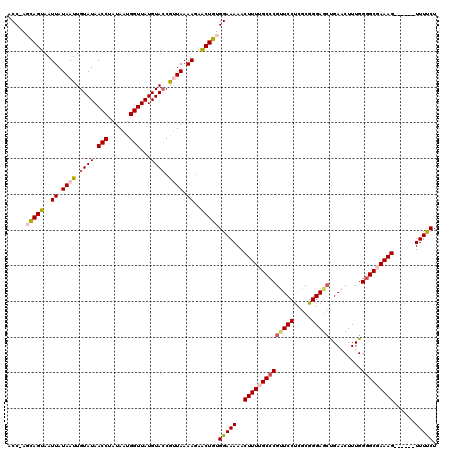
| Location | 8,500,479 – 8,500,593 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.87 |
| Mean single sequence MFE | -34.64 |
| Consensus MFE | -26.08 |
| Energy contribution | -27.00 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.973851 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8500479 114 + 22407834 ACCAAGCAGUAAUUAUAAUUGUAUAACCUAUAAUGGUUAUGUAGCGUUAAAAGAACUGUGGAAAAACUUUUGCCCGCUCCUCGCGGGAGCUGAACUUUGGGGCGAAAG------UUUUCU .((..(((((..((.(((((((((((((......)))))))))..)))).))..))))))).((((((((((((((((((.....))))).........)))))))))------)))).. ( -38.10) >DroSec_CAF1 9916 113 + 1 ACC-AGCAGUAAUUAUAAUUGUAUAACCUAUAAUGGUUAUGUACCGCUAAAAUAACUGUGGAAAAACUUUUGCCCGUUCCUCGCGGGAGCUGAACUUUGGGGCGAAAG------UUUUCU .((-(.((((.(((.((..(((((((((......)))))))))....)).))).))))))).(((((((((((((((((...((....)).))))....)))))))))------)))).. ( -35.50) >DroSim_CAF1 9125 113 + 1 ACC-AGCAGUAAUUAUAAUUGUAUAACCUAUAAUGGUUAUGUACCGCUAAAAGAACUGUGGAAAAACUUUUGCCCGUUCCUCGCGGGAGCUGAACUUUGGGGCGAAAG------UUUUCU .((-(.((((..((.((..(((((((((......)))))))))....)).))..))))))).(((((((((((((((((...((....)).))))....)))))))))------)))).. ( -34.60) >DroEre_CAF1 9535 119 + 1 ACC-ACUAGUAAUUAUAAUUGUAUAACCUAUAAUGGUUAUGUACCAUUAAAAGAGCUGUGGAAAAACUUUAGCCCGUUCCCCGCGGGAGGCGAACUUUGGGGCGAAAGCGAAAGUUUCCU .((-((.(((..((.((((.((((((((......)))..))))).)))).))..)))))))..(((((((..(((((((.((......)).))))...)))((....)).)))))))... ( -34.70) >DroYak_CAF1 10115 114 + 1 ACC-ACUAGUAAUUAUAAUUGUAUAACCUAUAAUGGUUAUGUACCAUUAAAAGAGCUGUGGAAAAACUUUGGCCCGUUCCUCGCGGGAUCUGAACUUUGGAGCGAAAGU-----UUUCCU .((-((.(((..((.((((.((((((((......)))..))))).)))).))..))))))).((((((((.((((((.....))))..((..(...)..)))).)))))-----)))... ( -30.30) >consensus ACC_AGCAGUAAUUAUAAUUGUAUAACCUAUAAUGGUUAUGUACCGUUAAAAGAACUGUGGAAAAACUUUUGCCCGUUCCUCGCGGGAGCUGAACUUUGGGGCGAAAG______UUUUCU .....(((((..((.((((.((((((((......)))..))))).)))).))..)))))(((((..((((((((((((((.....))))).........)))))))))......))))). (-26.08 = -27.00 + 0.92)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:40:40 2006