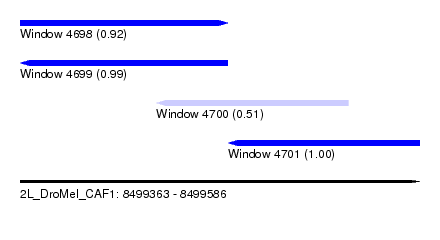
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,499,363 – 8,499,586 |
| Length | 223 |
| Max. P | 0.995176 |

| Location | 8,499,363 – 8,499,479 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.26 |
| Mean single sequence MFE | -29.29 |
| Consensus MFE | -22.36 |
| Energy contribution | -22.28 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.920818 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8499363 116 + 22407834 AGCAAAGCCUGCUAACGGCAGGAAAAGCCAUAUAAUUAGGGGAAAUUAUUGCCACGACAGG----CACUUUUAGUUGCAUUAAAUUUUCAUAUACCAAAUGAAAAUUGCGUAUACGCAUG .((((..(((((.....)))))(((((((.((....)).))........((((......))----)))))))..))))....(((((((((.......)))))))))(((....)))... ( -29.10) >DroSec_CAF1 8802 116 + 1 AGCAAAGCCUGCUAAUGGCAGGAAAAGCCAUAUAAUUAGGGGAAAUUAUUGCCGCGACAGG----CACUUUUAGUUGCAUUAAAUUUUCAUAUACCAAAUGAAAAUUGCGUAUACGCAUG .((((..(((((.....)))))(((((((.((....)).))........((((......))----)))))))..))))....(((((((((.......)))))))))(((....)))... ( -29.80) >DroSim_CAF1 8019 116 + 1 AGCAAAGCCUGCUAACGGCAGGAAAAGCCAUAUAAUUAGGGGAAAUUAUUGCCACGACAGG----CACUUUUAGUUGCAUUAAAUUUUCAUAUACCAAAUGAAAAUUGCGUAUACGCAUG .((((..(((((.....)))))(((((((.((....)).))........((((......))----)))))))..))))....(((((((((.......)))))))))(((....)))... ( -29.10) >DroEre_CAF1 8539 116 + 1 AGCAAAGCCUGCCAGUGGAAGGGAAAGCUAAAUAAUUAGGGCAAAUUAUUGCCACGGCAGC----CACUUUCAGUUGCAUUAAAUUUUCAUAUACCAAAUGAAAAUUGCGUAUACGCACG .((((...(((..(((((...(.....((((....))))(((((....)))))....)..)----))))..)))))))....(((((((((.......)))))))))(((....)))... ( -29.20) >DroYak_CAF1 8966 120 + 1 AGCAAAGCCCGCUAACCGCAGGGAAAGCAAAAUAAUUAGGGCAAAUUAUUGCCACGACAGCUUUCCACUUUCAGUUGCAUUAAAUUUUCAUAUACCAAAUGAAAAUUGCGUAUACGCAUG .((((.....((.....))..(((((((...........(((((....)))))......)))))))........))))....(((((((((.......)))))))))(((....)))... ( -29.23) >consensus AGCAAAGCCUGCUAACGGCAGGAAAAGCCAUAUAAUUAGGGGAAAUUAUUGCCACGACAGG____CACUUUUAGUUGCAUUAAAUUUUCAUAUACCAAAUGAAAAUUGCGUAUACGCAUG .((((..(((((.....)))))(((((...........(((.((....)).)).)............)))))..))))....(((((((((.......)))))))))(((....)))... (-22.36 = -22.28 + -0.08)

| Location | 8,499,363 – 8,499,479 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.26 |
| Mean single sequence MFE | -29.48 |
| Consensus MFE | -25.56 |
| Energy contribution | -24.96 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.16 |
| SVM RNA-class probability | 0.989394 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8499363 116 - 22407834 CAUGCGUAUACGCAAUUUUCAUUUGGUAUAUGAAAAUUUAAUGCAACUAAAAGUG----CCUGUCGUGGCAAUAAUUUCCCCUAAUUAUAUGGCUUUUCCUGCCGUUAGCAGGCUUUGCU ..((((....))))((((((((.......)))))))).....((((..(((((((----((......))).((((((......))))))...))))))(((((.....)))))..)))). ( -28.30) >DroSec_CAF1 8802 116 - 1 CAUGCGUAUACGCAAUUUUCAUUUGGUAUAUGAAAAUUUAAUGCAACUAAAAGUG----CCUGUCGCGGCAAUAAUUUCCCCUAAUUAUAUGGCUUUUCCUGCCAUUAGCAGGCUUUGCU ..((((....))))((((((((.......)))))))).....((((..(((((((----((......))).((((((......))))))...))))))(((((.....)))))..)))). ( -27.70) >DroSim_CAF1 8019 116 - 1 CAUGCGUAUACGCAAUUUUCAUUUGGUAUAUGAAAAUUUAAUGCAACUAAAAGUG----CCUGUCGUGGCAAUAAUUUCCCCUAAUUAUAUGGCUUUUCCUGCCGUUAGCAGGCUUUGCU ..((((....))))((((((((.......)))))))).....((((..(((((((----((......))).((((((......))))))...))))))(((((.....)))))..)))). ( -28.30) >DroEre_CAF1 8539 116 - 1 CGUGCGUAUACGCAAUUUUCAUUUGGUAUAUGAAAAUUUAAUGCAACUGAAAGUG----GCUGCCGUGGCAAUAAUUUGCCCUAAUUAUUUAGCUUUCCCUUCCACUGGCAGGCUUUGCU ..((((....))))((((((((.......)))))))).....(((((((..((((----(.......(((((....)))))((((....)))).........)))))..)))...)))). ( -29.40) >DroYak_CAF1 8966 120 - 1 CAUGCGUAUACGCAAUUUUCAUUUGGUAUAUGAAAAUUUAAUGCAACUGAAAGUGGAAAGCUGUCGUGGCAAUAAUUUGCCCUAAUUAUUUUGCUUUCCCUGCGGUUAGCGGGCUUUGCU ..((((....))))((((((((.......)))))))).....((((..(((((..(((.((....))(((((....))))).......)))..)))))(((((.....)))))..)))). ( -33.70) >consensus CAUGCGUAUACGCAAUUUUCAUUUGGUAUAUGAAAAUUUAAUGCAACUAAAAGUG____CCUGUCGUGGCAAUAAUUUCCCCUAAUUAUAUGGCUUUUCCUGCCGUUAGCAGGCUUUGCU ..((((....))))((((((((.......)))))))).....((((..((((((.......(((....)))((((((......))))))...))))))(((((.....)))))..)))). (-25.56 = -24.96 + -0.60)

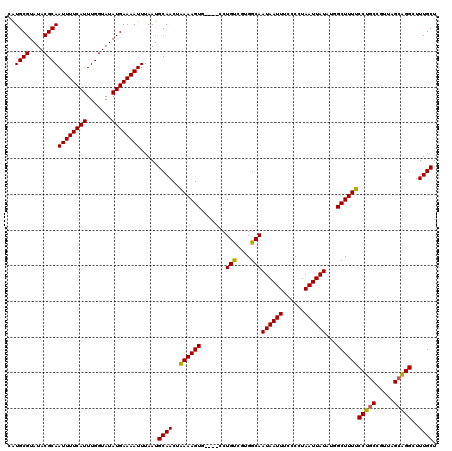
| Location | 8,499,439 – 8,499,546 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 81.67 |
| Mean single sequence MFE | -20.24 |
| Consensus MFE | -13.49 |
| Energy contribution | -13.49 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.67 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.505651 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8499439 107 - 22407834 AUUACAACGCGGCGGGUGUGGCGCUCUCU-UCCCCU-AC----CCCCUUCCCCAUCCAGUGCCAACCCCCUUCCAUGCGUAUACGCAAUUUUCAUUUGGUAUAUGAAAAUUUA ..........((.((((.(((((((....-......-..----..............))))))))))))).....((((....))))((((((((.......))))))))... ( -26.57) >DroSec_CAF1 8878 87 - 1 AUUACAACGCGGCGGGUGUGGCGCU--CU-UCCCCU-UC----CCCCU------------------CACCUUCCAUGCGUAUACGCAAUUUUCAUUUGGUAUAUGAAAAUUUA ..........((.(((((.((....--..-......-..----..)).------------------))))).)).((((....))))((((((((.......))))))))... ( -20.19) >DroSim_CAF1 8095 87 - 1 AUUACAACGCGGCGGGUGUGGCGCU--UU-UCCCCU-UC----CCCCU------------------CCCCUUCCAUGCGUAUACGCAAUUUUCAUUUGGUAUAUGAAAAUUUA ..........((.(((...((.(..--..-..))).-.)----)))).------------------.........((((....))))((((((((.......))))))))... ( -17.10) >DroYak_CAF1 9046 93 - 1 AUUACAACGCGGCGGGUGUGGUACU--CCCUCCCUUCCCCUCCUCCUC------------------CCCCUUCCAUGCGUAUACGCAAUUUUCAUUUGGUAUAUGAAAAUUUA ..........((.(((.(.((....--....))...)))).)).....------------------.........((((....))))((((((((.......))))))))... ( -17.10) >consensus AUUACAACGCGGCGGGUGUGGCGCU__CU_UCCCCU_UC____CCCCU__________________CCCCUUCCAUGCGUAUACGCAAUUUUCAUUUGGUAUAUGAAAAUUUA ........(((..(.((((((...................................................)))))).)...)))(((((((((.......))))))))).. (-13.49 = -13.49 + 0.00)

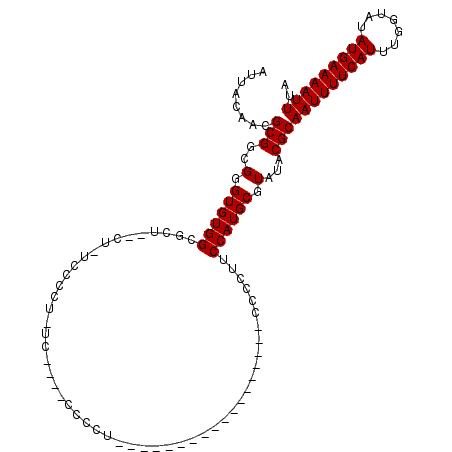
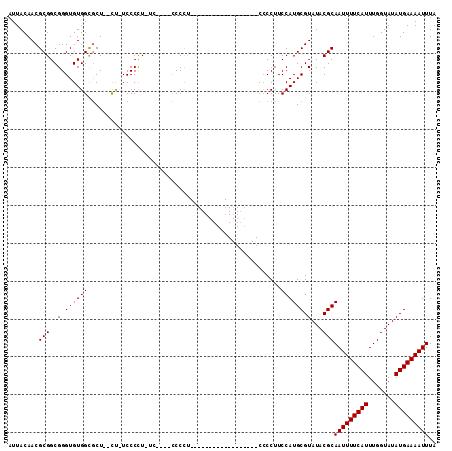
| Location | 8,499,479 – 8,499,586 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 81.17 |
| Mean single sequence MFE | -20.70 |
| Consensus MFE | -19.91 |
| Energy contribution | -20.16 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.55 |
| SVM RNA-class probability | 0.995176 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8499479 107 - 22407834 GCCAAUCCGCCGACUUUGUCACUAAUUCAAUAAGCAUCCAAUUACAACGCGGCGGGUGUGGCGCUCUCU-UCCCCU-AC----CCCCUUCCCCAUCCAGUGCCAACCCCCUUC ....((((((((.(.((((........................)))).))))))))).(((((((....-......-..----..............)))))))......... ( -22.33) >DroSec_CAF1 8918 87 - 1 GCCAAUCCGCCGACUUUGUCACUAAUUCAAUAAGCAUCCAAUUACAACGCGGCGGGUGUGGCGCU--CU-UCCCCU-UC----CCCCU------------------CACCUUC ((((((((((((.(.((((........................)))).))))))))).))))...--..-......-..----.....------------------....... ( -21.56) >DroSim_CAF1 8135 87 - 1 GCCAAUCCGCCGACUUUGUCACUAAUUCAAUAAGCAUCCAAUUACAACGCGGCGGGUGUGGCGCU--UU-UCCCCU-UC----CCCCU------------------CCCCUUC ((((((((((((.(.((((........................)))).))))))))).))))...--..-......-..----.....------------------....... ( -21.56) >DroYak_CAF1 9086 93 - 1 CCCAAUCCGCCGACUUUGUCACUAAUUCAAUAAGCAUCCAAUUACAACGCGGCGGGUGUGGUACU--CCCUCCCUUCCCCUCCUCCUC------------------CCCCUUC .(((((((((((.(.((((........................)))).))))))))).)))....--.....................------------------....... ( -17.36) >consensus GCCAAUCCGCCGACUUUGUCACUAAUUCAAUAAGCAUCCAAUUACAACGCGGCGGGUGUGGCGCU__CU_UCCCCU_UC____CCCCU__________________CCCCUUC ((((((((((((.(.((((........................)))).))))))))).))))................................................... (-19.91 = -20.16 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:40:38 2006