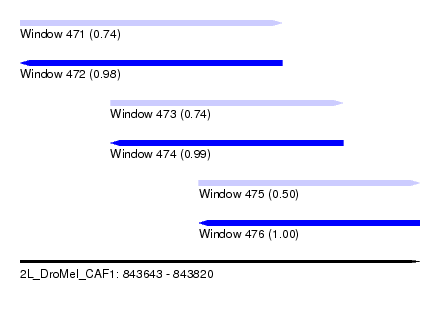
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 843,643 – 843,820 |
| Length | 177 |
| Max. P | 0.999518 |

| Location | 843,643 – 843,759 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 85.82 |
| Mean single sequence MFE | -32.67 |
| Consensus MFE | -20.03 |
| Energy contribution | -20.63 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.744298 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
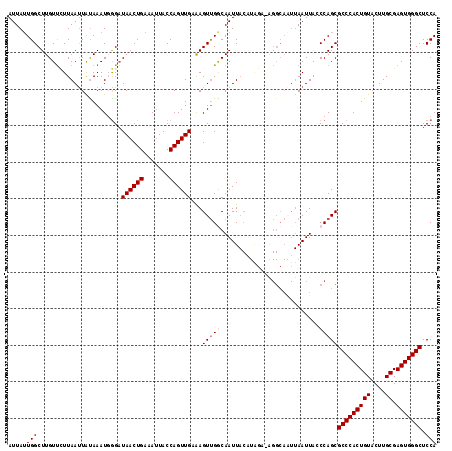
>2L_DroMel_CAF1 843643 116 + 22407834 AUUAUUGGCUUGUUAUUAAUUAUAAAUGGGAUAACUGAAAUUACCAGUUGAAAGUUGGCAAUUACAUAUG-AGGCAAUUAAUUACCCAGCGCCCACUGUACUUGCGAGUGGGCUCCA .....(((..................((((.((((((.......))))))..(((((.(...........-.).))))).....))))..(((((((((....)).))))))).))) ( -27.80) >DroSec_CAF1 9060 116 + 1 AUUAUUGGCUUGUUCUUAAUUACAAAUGGGAUAACUGACAAUACCAGUUGAAAGUUGGCAAUUACAUAGA-AGGCAAUUAAUUAGCCAGCGCCCACUGUACUUGCGAGUGGGCUCCA ......((((..(((.(((.((((..((((.((((((.......))))))...(((((((((((......-.......))))).)))))).)))).)))).))).)))..))))... ( -33.42) >DroSim_CAF1 9716 116 + 1 AUUAUUGGCUUGUUCUUAAUUAUAAAUGGGAUAACUGACAUUACCAGUUGAAAGUUGGCAAUUACAUAGA-AGGCAAUUAAUUAGCCAGCGCCCACUGUACUUGCGAGUGGGCUCCA ......((((..(((.(((.((((..((((.((((((.......))))))...(((((((((((......-.......))))).)))))).)))).)))).))).)))..))))... ( -31.52) >DroEre_CAF1 8612 101 + 1 AUUAAUGGCUUGUCUUUAACUAUAAAUUAGAUAACUGAAAUUACCAGUUGCGAGUCGACAAUU----------------AAUUACCCAGCGCCCACUGUACUUGCGAGUGGGCUCCA .....((((((((......(((.....)))..(((((.......)))))))))))))......----------------...........(((((((((....)).))))))).... ( -25.90) >DroYak_CAF1 7884 117 + 1 AUUAUUGGCUUGUUCUUAAUUGUAAAUUGGCUAACUGAAAUUACCAGUUGCAAGUUGACAAUUACUUGGCUGGGCAUUUAAUUGCCCAGCGCCCACUGUACUUGCGAGUGGGCUCCA .....(((........(((((((.((((.((.(((((.......))))))).)))).)))))))....((((((((......))))))))(((((((((....)).))))))).))) ( -44.70) >consensus AUUAUUGGCUUGUUCUUAAUUAUAAAUGGGAUAACUGAAAUUACCAGUUGAAAGUUGGCAAUUACAUAGA_AGGCAAUUAAUUACCCAGCGCCCACUGUACUUGCGAGUGGGCUCCA .....(((.......................((((((.......))))))...(((((...........................)))))(((((((((....)).))))))).))) (-20.03 = -20.63 + 0.60)

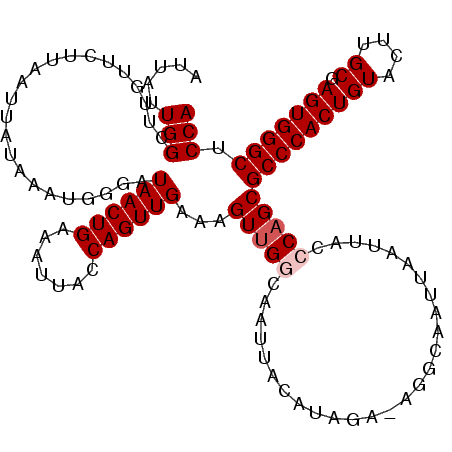
| Location | 843,643 – 843,759 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 85.82 |
| Mean single sequence MFE | -31.97 |
| Consensus MFE | -23.45 |
| Energy contribution | -23.64 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.94 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.95 |
| SVM RNA-class probability | 0.983624 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 843643 116 - 22407834 UGGAGCCCACUCGCAAGUACAGUGGGCGCUGGGUAAUUAAUUGCCU-CAUAUGUAAUUGCCAACUUUCAACUGGUAAUUUCAGUUAUCCCAUUUAUAAUUAAUAACAAGCCAAUAAU (((.((((((((....)...)))))))..((((....((((((...-.......((((((((.........)))))))).)))))).))))..................)))..... ( -29.50) >DroSec_CAF1 9060 116 - 1 UGGAGCCCACUCGCAAGUACAGUGGGCGCUGGCUAAUUAAUUGCCU-UCUAUGUAAUUGCCAACUUUCAACUGGUAUUGUCAGUUAUCCCAUUUGUAAUUAAGAACAAGCCAAUAAU ....((((((((....)...)))))))..(((((..((((((((..-.....(((((((.(((...((....))..))).))))))).......)))))))).....)))))..... ( -31.54) >DroSim_CAF1 9716 116 - 1 UGGAGCCCACUCGCAAGUACAGUGGGCGCUGGCUAAUUAAUUGCCU-UCUAUGUAAUUGCCAACUUUCAACUGGUAAUGUCAGUUAUCCCAUUUAUAAUUAAGAACAAGCCAAUAAU .(((((((((((....)...)))))))((((((........(((..-.....)))(((((((.........)))))))))))))..)))............................ ( -28.30) >DroEre_CAF1 8612 101 - 1 UGGAGCCCACUCGCAAGUACAGUGGGCGCUGGGUAAUU----------------AAUUGUCGACUCGCAACUGGUAAUUUCAGUUAUCUAAUUUAUAGUUAAAGACAAGCCAUUAAU ....((((((((....)...)))))))(((..((..((----------------((((((.((.....((((((.....))))))......)).))))))))..)).)))....... ( -27.00) >DroYak_CAF1 7884 117 - 1 UGGAGCCCACUCGCAAGUACAGUGGGCGCUGGGCAAUUAAAUGCCCAGCCAAGUAAUUGUCAACUUGCAACUGGUAAUUUCAGUUAGCCAAUUUACAAUUAAGAACAAGCCAAUAAU (((.((((((((....)...)))))))((((((((......))))))))....(((((((......((((((((.....)))))).))......)))))))........)))..... ( -43.50) >consensus UGGAGCCCACUCGCAAGUACAGUGGGCGCUGGGUAAUUAAUUGCCU_UCUAUGUAAUUGCCAACUUUCAACUGGUAAUUUCAGUUAUCCCAUUUAUAAUUAAGAACAAGCCAAUAAU ....((((((((....)...)))))))(((.((((((((..((......))..)))))))).......((((((.....))))))......................)))....... (-23.45 = -23.64 + 0.19)

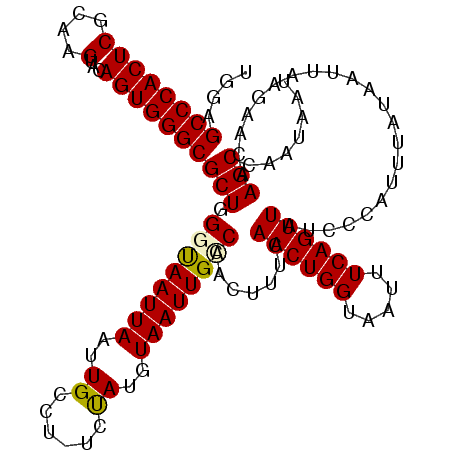
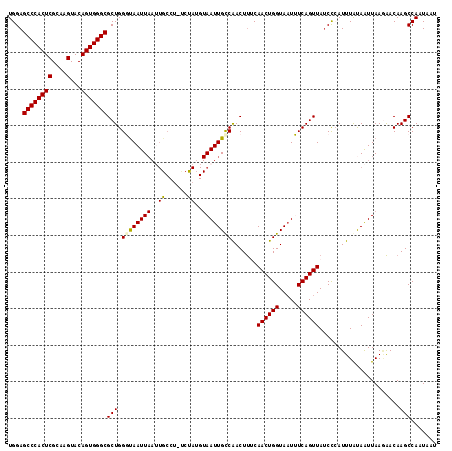
| Location | 843,683 – 843,786 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 78.78 |
| Mean single sequence MFE | -28.89 |
| Consensus MFE | -17.67 |
| Energy contribution | -18.27 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.743344 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 843683 103 + 22407834 UUACCAGUUGAAAGUUGGCAAUUACAUAUG-AGGCAAUUAAUUACCCAGCGCCCACUGUACUUGCGAGUGGGCUCCACAAAUAUUGGGCGAGUG----UUUGUUUGGC ...((((......(((((.(((((......-.......)))))..)))))(((((((((....)).)))))))...(((((((((.....))))----))))))))). ( -26.32) >DroSec_CAF1 9100 92 + 1 AUACCAGUUGAAAGUUGGCAAUUACAUAGA-AGGCAAUUAAUUAGCCAGCGCCCACUGUACUUGCGAGUGGGCUCCACAAAUAU---------------UUGUUUGGC ...((((..(((.(((((((((((......-.......))))).))))))(((((((((....)).)))))))..........)---------------))..)))). ( -26.52) >DroSim_CAF1 9756 92 + 1 UUACCAGUUGAAAGUUGGCAAUUACAUAGA-AGGCAAUUAAUUAGCCAGCGCCCACUGUACUUGCGAGUGGGCUCCACAAAUAU---------------UUGUUUGGC ...((((..(((.(((((((((((......-.......))))).))))))(((((((((....)).)))))))..........)---------------))..)))). ( -26.52) >DroEre_CAF1 8652 92 + 1 UUACCAGUUGCGAGUCGACAAUU----------------AAUUACCCAGCGCCCACUGUACUUGCGAGUGGGCUCCACAAAUAUUAGGUGAAUGUCUCUCUGUUUGGC ...((((..(((((..((((...----------------...........(((((((((....)).)))))))..(((.........)))..)))).))).)))))). ( -24.00) >DroYak_CAF1 7924 108 + 1 UUACCAGUUGCAAGUUGACAAUUACUUGGCUGGGCAUUUAAUUGCCCAGCGCCCACUGUACUUGCGAGUGGGCUCCACAAAUAUUGGCUGAAUAUCUGUUUGUUUGGC ....((((..(((((........)))))((((((((......))))))))(((((((((....)).))))))).........))))((..((((......))))..)) ( -41.10) >consensus UUACCAGUUGAAAGUUGGCAAUUACAUAGA_AGGCAAUUAAUUACCCAGCGCCCACUGUACUUGCGAGUGGGCUCCACAAAUAUU_G__GA_U_____UUUGUUUGGC ...((((......(((((...........................)))))(((((((((....)).)))))))..............................)))). (-17.67 = -18.27 + 0.60)

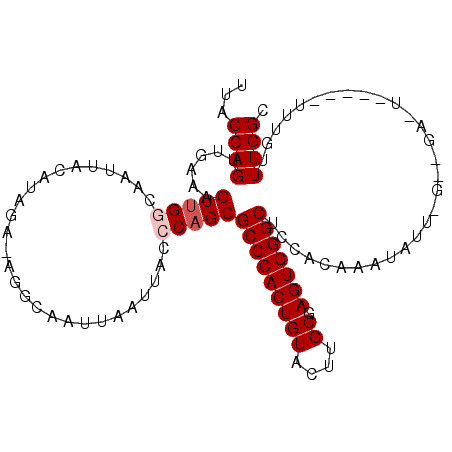
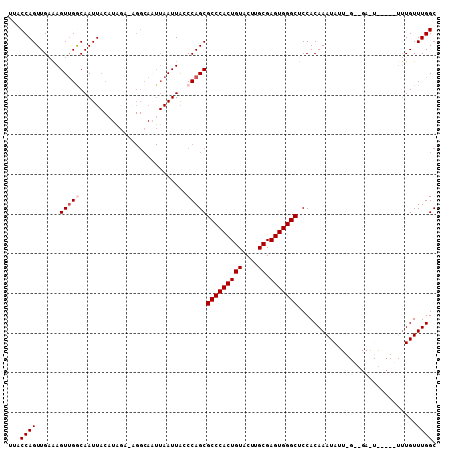
| Location | 843,683 – 843,786 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 78.78 |
| Mean single sequence MFE | -29.62 |
| Consensus MFE | -21.03 |
| Energy contribution | -21.22 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.71 |
| SVM decision value | 2.49 |
| SVM RNA-class probability | 0.994537 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 843683 103 - 22407834 GCCAAACAAA----CACUCGCCCAAUAUUUGUGGAGCCCACUCGCAAGUACAGUGGGCGCUGGGUAAUUAAUUGCCU-CAUAUGUAAUUGCCAACUUUCAACUGGUAA ((((......----((..((((((.((((((((.((....))))))))))...)))))).))((((((((..((...-))....))))))))..........)))).. ( -29.20) >DroSec_CAF1 9100 92 - 1 GCCAAACAA---------------AUAUUUGUGGAGCCCACUCGCAAGUACAGUGGGCGCUGGCUAAUUAAUUGCCU-UCUAUGUAAUUGCCAACUUUCAACUGGUAU ((((.((((---------------....))))(((((((((((....)...)))))))..((((.....((((((..-.....))))))))))...)))...)))).. ( -24.90) >DroSim_CAF1 9756 92 - 1 GCCAAACAA---------------AUAUUUGUGGAGCCCACUCGCAAGUACAGUGGGCGCUGGCUAAUUAAUUGCCU-UCUAUGUAAUUGCCAACUUUCAACUGGUAA ((((.((((---------------....))))(((((((((((....)...)))))))..((((.....((((((..-.....))))))))))...)))...)))).. ( -24.90) >DroEre_CAF1 8652 92 - 1 GCCAAACAGAGAGACAUUCACCUAAUAUUUGUGGAGCCCACUCGCAAGUACAGUGGGCGCUGGGUAAUU----------------AAUUGUCGACUCGCAACUGGUAA ((((....(((.(((((((((.........)))))((((((((....)...)))))))...........----------------...))))..))).....)))).. ( -27.60) >DroYak_CAF1 7924 108 - 1 GCCAAACAAACAGAUAUUCAGCCAAUAUUUGUGGAGCCCACUCGCAAGUACAGUGGGCGCUGGGCAAUUAAAUGCCCAGCCAAGUAAUUGUCAACUUGCAACUGGUAA ((((.....(((((((((.....)))))))))...((((((((....)...)))))))((((((((......))))))))(((((........)))))....)))).. ( -41.50) >consensus GCCAAACAAA_____A_UC__C_AAUAUUUGUGGAGCCCACUCGCAAGUACAGUGGGCGCUGGGUAAUUAAUUGCCU_UCUAUGUAAUUGCCAACUUUCAACUGGUAA ((((............................((.((((((((....)...))))))).)).((((((((..((......))..))))))))..........)))).. (-21.03 = -21.22 + 0.19)

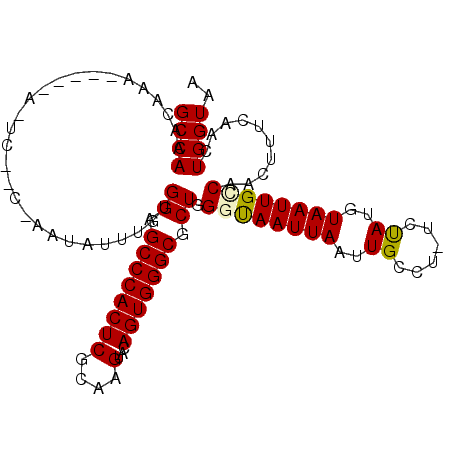
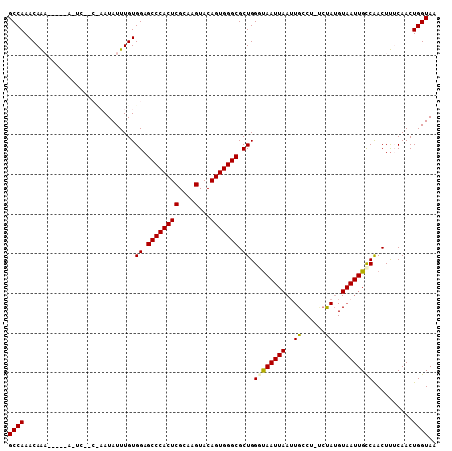
| Location | 843,722 – 843,820 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 87.86 |
| Mean single sequence MFE | -25.76 |
| Consensus MFE | -22.10 |
| Energy contribution | -22.10 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.86 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 843722 98 + 22407834 AUUACCCAGCGCCCACUGUACUUGCGAGUGGGCUCCACAAAUAUUGGGCGAGUG----UUUGUUUGGCAAAUAGAUGAAAAUUGCCGCUAACUUUGAUUUGG .......((((((((((((....)).))))))).............(((((...----((..((((.....))))..))..))))))))............. ( -28.20) >DroSec_CAF1 9139 87 + 1 AUUAGCCAGCGCCCACUGUACUUGCGAGUGGGCUCCACAAAUAU---------------UUGUUUGGCAAAUAGAUGAAAAUUGCCGCUAACUUUGAUUUGG .(((((..(((((((((((....)).)))))))....((..(((---------------(((.....))))))..))......)).)))))........... ( -25.00) >DroSim_CAF1 9795 87 + 1 AUUAGCCAGCGCCCACUGUACUUGCGAGUGGGCUCCACAAAUAU---------------UUGUUUGGCAAAUAGAUGAAAAUUGCCGCUAACUUUGAUUUGG .(((((..(((((((((((....)).)))))))....((..(((---------------(((.....))))))..))......)).)))))........... ( -25.00) >DroEre_CAF1 8676 102 + 1 AUUACCCAGCGCCCACUGUACUUGCGAGUGGGCUCCACAAAUAUUAGGUGAAUGUCUCUCUGUUUGGCAAAUAAAUGAAAAUUGCCGCUAACUUUGAUUUGG ..........(((((((((....)).)))))))....(((((...((((((((........))))(((((...........)))))....))))..))))). ( -23.50) >DroYak_CAF1 7964 102 + 1 AUUGCCCAGCGCCCACUGUACUUGCGAGUGGGCUCCACAAAUAUUGGCUGAAUAUCUGUUUGUUUGGCAAAUAAAUGAAAAUUGCCGCUAACUUUGAUUUGG .....((((.(((((((((....)).)))))))....((((..(((((.(((((......)))))(((((...........)))))))))).))))..)))) ( -27.10) >consensus AUUACCCAGCGCCCACUGUACUUGCGAGUGGGCUCCACAAAUAUU_G__GA_U_____UUUGUUUGGCAAAUAGAUGAAAAUUGCCGCUAACUUUGAUUUGG .......((((((((((((....)).)))))))................................(((((...........))))))))............. (-22.10 = -22.10 + -0.00)

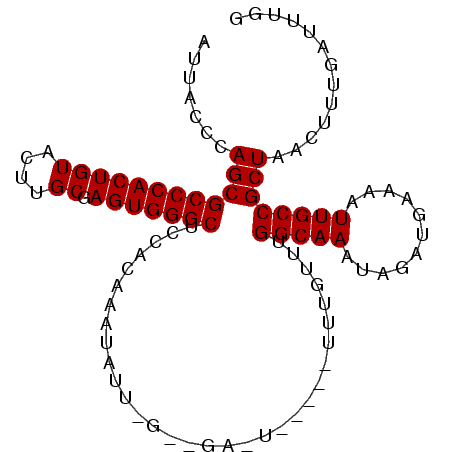
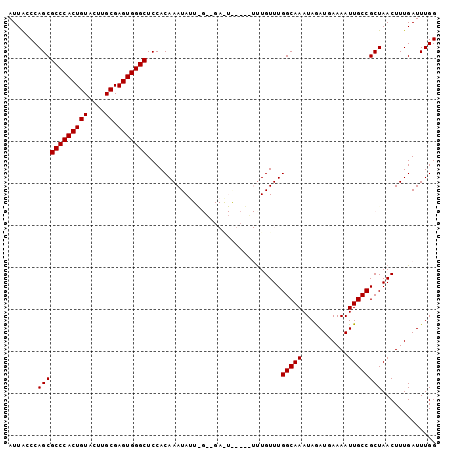
| Location | 843,722 – 843,820 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 87.86 |
| Mean single sequence MFE | -29.38 |
| Consensus MFE | -25.18 |
| Energy contribution | -25.18 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.16 |
| Structure conservation index | 0.86 |
| SVM decision value | 3.68 |
| SVM RNA-class probability | 0.999518 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 843722 98 - 22407834 CCAAAUCAAAGUUAGCGGCAAUUUUCAUCUAUUUGCCAAACAAA----CACUCGCCCAAUAUUUGUGGAGCCCACUCGCAAGUACAGUGGGCGCUGGGUAAU .........(.((((((((((...........))))).......----.....(((((.((((((((.((....))))))))))...)))))))))).)... ( -29.00) >DroSec_CAF1 9139 87 - 1 CCAAAUCAAAGUUAGCGGCAAUUUUCAUCUAUUUGCCAAACAA---------------AUAUUUGUGGAGCCCACUCGCAAGUACAGUGGGCGCUGGCUAAU .........((((((((((((...........)))))......---------------...........((((((((....)...))))))))))))))... ( -29.90) >DroSim_CAF1 9795 87 - 1 CCAAAUCAAAGUUAGCGGCAAUUUUCAUCUAUUUGCCAAACAA---------------AUAUUUGUGGAGCCCACUCGCAAGUACAGUGGGCGCUGGCUAAU .........((((((((((((...........)))))......---------------...........((((((((....)...))))))))))))))... ( -29.90) >DroEre_CAF1 8676 102 - 1 CCAAAUCAAAGUUAGCGGCAAUUUUCAUUUAUUUGCCAAACAGAGAGACAUUCACCUAAUAUUUGUGGAGCCCACUCGCAAGUACAGUGGGCGCUGGGUAAU .........(.((((((((((...........)))))..(((((.((........))....)))))...((((((((....)...)))))))))))).)... ( -27.50) >DroYak_CAF1 7964 102 - 1 CCAAAUCAAAGUUAGCGGCAAUUUUCAUUUAUUUGCCAAACAAACAGAUAUUCAGCCAAUAUUUGUGGAGCCCACUCGCAAGUACAGUGGGCGCUGGGCAAU ..........(((((((((((...........)))))......(((((((((.....)))))))))...((((((((....)...)))))))))).)))... ( -30.60) >consensus CCAAAUCAAAGUUAGCGGCAAUUUUCAUCUAUUUGCCAAACAAA_____A_UC__C_AAUAUUUGUGGAGCCCACUCGCAAGUACAGUGGGCGCUGGGUAAU ...........((((((((((...........)))))................................((((((((....)...))))))))))))..... (-25.18 = -25.18 + -0.00)

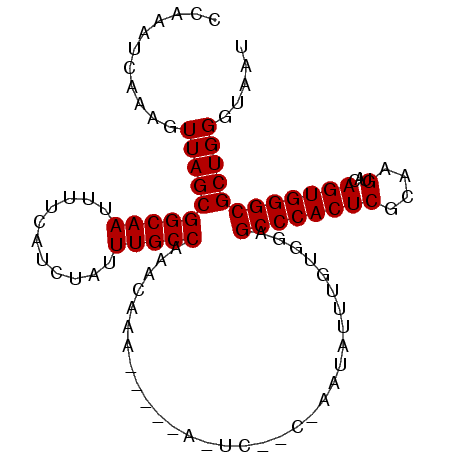
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:32:23 2006