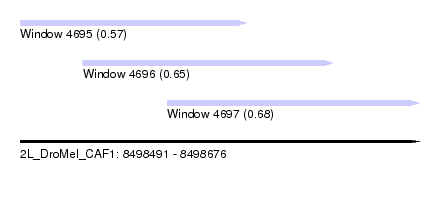
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,498,491 – 8,498,676 |
| Length | 185 |
| Max. P | 0.680841 |

| Location | 8,498,491 – 8,498,596 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.26 |
| Mean single sequence MFE | -37.08 |
| Consensus MFE | -24.33 |
| Energy contribution | -25.59 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.567729 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
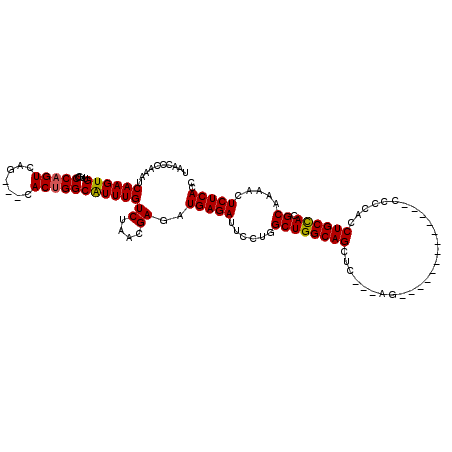
>2L_DroMel_CAF1 8498491 105 + 22407834 -----------CCGCAGUGGCCGCCGCAUUCAAAUCCCAUCGAGC-GGAAUAGGCCAAACAGUUUGGCCUGCUUGGUGGCUAACCCAAAUCAAGUGUUGCCCAGUCAG---CACUGGCAU -----------..((..((((((((((((((...((.....))..-.))))(((((((.....)))))))))..))))))))..........(((((((......)))---)))).)).. ( -37.20) >DroSim_CAF1 7155 105 + 1 -----------CCGCAGUGUCCGCAGCAUUCAAAUCCCAUCAAGC-GGAAUAGGCCAAACAGUUUGGGCUGCUUGGUGGCUAACCCAAAUCAAGUGUUGCCCAGUCAG---CACUGGCAU -----------..(((((((..((((((((......(((((((((-((..(((((......)))))..))))))))))).............)))))))).......)---)))).)).. ( -33.81) >DroEre_CAF1 7511 117 + 1 GAGUGGUCGGCAUGCCAUGGCCGCAGCAUUCAAAUCCCAUCAAAUGGGAAUUGGCCAAACAGCUUGGCCCGCUUGCUGGUUAACCCAAAUCAAGUGUUGCCCAGUCAG---CACAGGCGU (((((.(((((........)))).).)))))...((((((...))))))...((((((.....)))))).(((.(((((.((((.(.......).)))).))))).))---)........ ( -41.90) >DroYak_CAF1 7935 108 + 1 -----------AUUCCAUGGCCGCAGCAUUCAAAUCCCAUCAAGC-GGAAUAGGCCAAACAGUGUGGGCCGCUCGGUGGUUAACCCAAAUCAAGUGUUGCCCAGUCAGCAGCACUGGCGU -----------........(((..............(((((.(((-((..((.((......)).))..))))).))))).............((((((((.......))))))))))).. ( -35.40) >consensus ___________ACGCAAUGGCCGCAGCAUUCAAAUCCCAUCAAGC_GGAAUAGGCCAAACAGUUUGGCCCGCUUGGUGGCUAACCCAAAUCAAGUGUUGCCCAGUCAG___CACUGGCAU .................(((..((((((((......(((((((((.((..(((((......)))))..))))))))))).............)))))))))))(((((.....))))).. (-24.33 = -25.59 + 1.25)

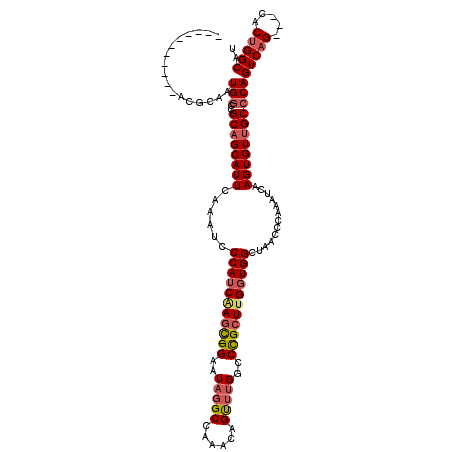
| Location | 8,498,520 – 8,498,636 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.52 |
| Mean single sequence MFE | -40.60 |
| Consensus MFE | -28.30 |
| Energy contribution | -29.05 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.654349 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8498520 116 + 22407834 CGAGC-GGAAUAGGCCAAACAGUUUGGCCUGCUUGGUGGCUAACCCAAAUCAAGUGUUGCCCAGUCAG---CACUGGCAUUUGUCUAACGAGAUGAGAUUCCUGGCUGGCAGCUCCACAA .((((-((...(((((((.....)))))))(((((((((.....)))..)))))).....)).(((((---(...((.((((((((....)))).)))).))..)))))).))))..... ( -41.10) >DroSim_CAF1 7184 116 + 1 CAAGC-GGAAUAGGCCAAACAGUUUGGGCUGCUUGGUGGCUAACCCAAAUCAAGUGUUGCCCAGUCAG---CACUGGCAUUUGUCUAACGAGAUGAGAUUUCUGGCUGGCAGCUCCACUG ..((.-(((....((((..(((..(((((.(((((((((.....)))..))))))...)))))((((.---...)))).((..(((....)))..))....)))..))))...))).)). ( -37.60) >DroEre_CAF1 7551 110 + 1 CAAAUGGGAAUUGGCCAAACAGCUUGGCCCGCUUGCUGGUUAACCCAAAUCAAGUGUUGCCCAGUCAG---CACAGGCGUUUGUCUAACGAGAUGAGAUUCCUGGCUGGCAGC------- ....((((....((((((.....))))))((((((.(((.....)))...))))))...))))(((((---(.((((..((..(((....)))..))...))))))))))...------- ( -43.20) >DroYak_CAF1 7964 116 + 1 CAAGC-GGAAUAGGCCAAACAGUGUGGGCCGCUCGGUGGUUAACCCAAAUCAAGUGUUGCCCAGUCAGCAGCACUGGCGUUUGUCUAACGAGAUGAGAUUCCUGGCUGGCAGCAC---AG ..(((-((..((.((......)).))..))))).(((.....)))........((((((((.((((((..((....)).((..(((....)))..))....))))))))))))))---.. ( -40.50) >consensus CAAGC_GGAAUAGGCCAAACAGUUUGGCCCGCUUGGUGGCUAACCCAAAUCAAGUGUUGCCCAGUCAG___CACUGGCAUUUGUCUAACGAGAUGAGAUUCCUGGCUGGCAGCUC___AG .............((((..(((..(((((((((((.(((.....)))...))))))..)))))(((((.....))))).((..(((....)))..))....)))..)))).......... (-28.30 = -29.05 + 0.75)

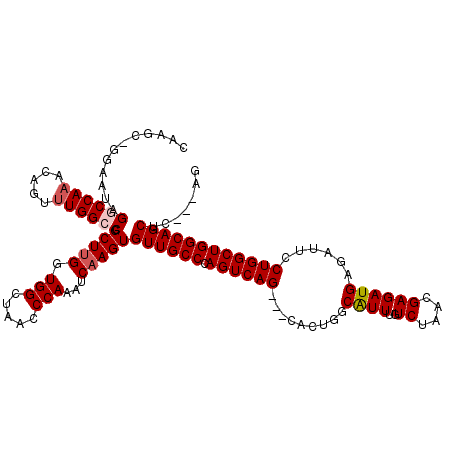

| Location | 8,498,559 – 8,498,676 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.04 |
| Mean single sequence MFE | -27.03 |
| Consensus MFE | -19.41 |
| Energy contribution | -19.41 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.680841 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8498559 117 + 22407834 UAACCCAAAUCAAGUGUUGCCCAGUCAG---CACUGGCAUUUGUCUAACGAGAUGAGAUUCCUGGCUGGCAGCUCCACAACCAUCCACCCCACACCCACCUGCUACGCAAAACUCUCACC .....((((((.(((((((......)))---)))).).)))))((....))..(((((......((((((((...........................)))))).)).....))))).. ( -22.93) >DroSim_CAF1 7223 111 + 1 UAACCCAAAUCAAGUGUUGCCCAGUCAG---CACUGGCAUUUGUCUAACGAGAUGAGAUUUCUGGCUGGCAGCUCCACUGCCA------CUGCCCCCACCUGCUACGCAAAACUCUCACC .....((((((.(((((((......)))---)))).).)))))((....))..(((((.....(((((((((.....))))))------..)))......(((...)))....))))).. ( -28.60) >DroEre_CAF1 7591 95 + 1 UAACCCAAAUCAAGUGUUGCCCAGUCAG---CACAGGCGUUUGUCUAACGAGAUGAGAUUCCUGGCUGGCAGC----------------------CCACCUGCCACGCAAAACUCUCACC ............(((.((((......((---(.((((..((..(((....)))..))...)))))))(((((.----------------------....)))))..)))).)))...... ( -26.80) >DroYak_CAF1 8003 104 + 1 UAACCCAAAUCAAGUGUUGCCCAGUCAGCAGCACUGGCGUUUGUCUAACGAGAUGAGAUUCCUGGCUGGCAGCAC---AG-------------CCCCACCUGCCACGCAAAACUCUCACC .....(((((..((((((((.......))))))))...)))))((....))..(((((......((((((((...---..-------------......)))))).)).....))))).. ( -29.80) >consensus UAACCCAAAUCAAGUGUUGCCCAGUCAG___CACUGGCAUUUGUCUAACGAGAUGAGAUUCCUGGCUGGCAGCUC___AG_____________CCCCACCUGCCACGCAAAACUCUCACC ........(((...(((((..(((((((.....)))))...))..))))).)))((((......((((((((...........................)))))).)).....))))... (-19.41 = -19.41 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:40:34 2006