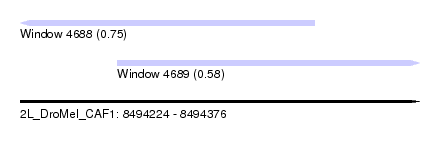
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,494,224 – 8,494,376 |
| Length | 152 |
| Max. P | 0.750082 |

| Location | 8,494,224 – 8,494,336 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 91.58 |
| Mean single sequence MFE | -34.96 |
| Consensus MFE | -26.74 |
| Energy contribution | -27.94 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.750082 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8494224 112 - 22407834 AGUGAAGCAGGAAGG--G-GGGGAGAAAACAGGAAGGGUAGCCCAGAUUGGGACUGCUUCGUUGAGCAGUUAAUGAAUCACUUAAUGGAAAAACCUGGACGCCACUCGACUUCCG-- .........(((((.--.-(((..(....((((....(((((((.....))).))))(((((((((..(((....)))..)))))))))....))))....)..)))..))))).-- ( -36.60) >DroSec_CAF1 3398 114 - 1 AGUGGAGCAGGAAGGAGG-GGGGAGAGAACAGGAAGGGGAGCCCAGAUUGGGACUGCUUCGUUGAGCAGUUAAUGAAUCACUUAAUGGAAAAACCUGGUCGCCACUCGACUUCCG-- .........(((((..((-(.((...((.((((..(((...))).((((..((((((((....))))))))....))))..............)))).)).)).)))..))))).-- ( -38.80) >DroSim_CAF1 3416 114 - 1 AGUGGGGCAGGAAGGAGG-GGGGAGAGAACAGGAAGGGUAGCCCAGAUUGGGACUGCUUCGUUGAGCAGUUAAUGAAUCACUUAAUGGAAAAACCUGGUCGCCACUCGACUUCCG-- .........(((((..((-(.((...((.((((....(((((((.....))).))))(((((((((..(((....)))..)))))))))....)))).)).)).)))..))))).-- ( -40.50) >DroEre_CAF1 3025 109 - 1 AGUUGAGCAGGAAGGUCG------GAAAACAGGAAGGGUAACCCAGAUUGGGACUGCUUCGUUGAGCAGUUAAUGAAUCACUUAAUGGAAAAACCUGGUCGCCACUCGACUUCCG-- (((((((......((.((------(....((((..(((...))).((((..((((((((....))))))))....))))..............)))).))))).)))))))....-- ( -30.60) >DroYak_CAF1 3405 117 - 1 AUUGAAGCAGAAAGGAUGGGGGAAGAAAACAGGAAGGGUAACCCAGAUUGGGAAUGCUUCGUUGAGCAGUUAAUGAAUCACUUAAUGGAAAAACCUGGUCGCCACUCGACUUCCGUU ..............((((((((..((.........(((...))).(((..((.....(((((((((..(((....)))..)))))))))....))..))).....))..)))))))) ( -28.30) >consensus AGUGGAGCAGGAAGGAGG_GGGGAGAAAACAGGAAGGGUAGCCCAGAUUGGGACUGCUUCGUUGAGCAGUUAAUGAAUCACUUAAUGGAAAAACCUGGUCGCCACUCGACUUCCG__ .........(((((.......((.((...((((..(((...))).((((..((((((((....))))))))....))))..............)))).)).))......)))))... (-26.74 = -27.94 + 1.20)


| Location | 8,494,261 – 8,494,376 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 90.37 |
| Mean single sequence MFE | -23.05 |
| Consensus MFE | -17.84 |
| Energy contribution | -18.36 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.582231 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
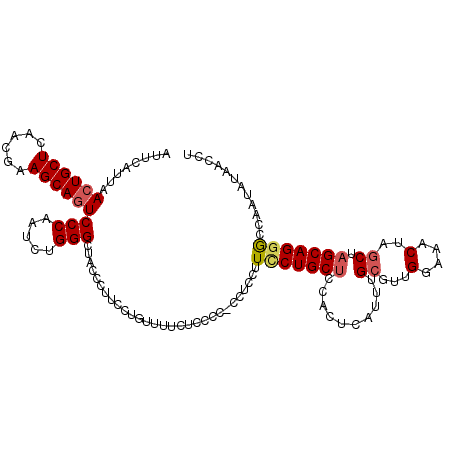
>2L_DroMel_CAF1 8494261 115 + 22407834 AUUCAUUAACUGCUCAACGAAGCAGUCCCAAUCUGGGCUACCCUUCCUGUUUUCUCCCC-C--CCUUCCUGCUUCACUCAUUUGGGUUGGAAACUAGCUAGCAGGGCCAAUAUAACCU ........((((((......)))))).........((((...((..(((.(((((..((-(--....................)))..))))).)))..))...)))).......... ( -23.85) >DroSec_CAF1 3435 117 + 1 AUUCAUUAACUGCUCAACGAAGCAGUCCCAAUCUGGGCUCCCCUUCCUGUUCUCUCCCC-CCUCCUUCCUGCUCCACUCAUUUGCGUUGGAAACUAGCUAGCAGGGCCAAUAUAACCU ........((((((......))))))(((.....)))......................-......(((((((..........((...(....)..)).)))))))............ ( -21.70) >DroSim_CAF1 3453 117 + 1 AUUCAUUAACUGCUCAACGAAGCAGUCCCAAUCUGGGCUACCCUUCCUGUUCUCUCCCC-CCUCCUUCCUGCCCCACUCAUUUGCGUUGGAAACUAGCUAGCAGGACCAAUAUAACCU ........((((((......))))))(((.....)))......................-......((((((...........((...(....)..))..))))))............ ( -21.02) >DroEre_CAF1 3062 112 + 1 AUUCAUUAACUGCUCAACGAAGCAGUCCCAAUCUGGGUUACCCUUCCUGUUUUC------CGACCUUCCUGCUCAACUCAUUUGCGUUGGAAACUAGCCAGCAGCGGCAAUGUAACCU ........((((((......))))))........(((((((.........((((------((((......((...........))))))))))...(((......)))...))))))) ( -26.20) >DroYak_CAF1 3444 118 + 1 AUUCAUUAACUGCUCAACGAAGCAUUCCCAAUCUGGGUUACCCUUCCUGUUUUCUUCCCCCAUCCUUUCUGCUUCAAUCAUUUGCGUUGGCAACUAGCUAGCAGGGCCAAUAUAACCU ...........((((...((((((.........((((.....................)))).......))))))..........((((((.....)))))).))))........... ( -22.49) >consensus AUUCAUUAACUGCUCAACGAAGCAGUCCCAAUCUGGGCUACCCUUCCUGUUUUCUCCCC_CCUCCUUCCUGCUCCACUCAUUUGCGUUGGAAACUAGCUAGCAGGGCCAAUAUAACCU ........((((((......))))))(((.....))).............................(((((((..........((...(....)..)).)))))))............ (-17.84 = -18.36 + 0.52)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:40:26 2006