
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,483,709 – 8,483,829 |
| Length | 120 |
| Max. P | 0.805943 |

| Location | 8,483,709 – 8,483,807 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 88.64 |
| Mean single sequence MFE | -42.33 |
| Consensus MFE | -35.35 |
| Energy contribution | -35.00 |
| Covariance contribution | -0.35 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.805943 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
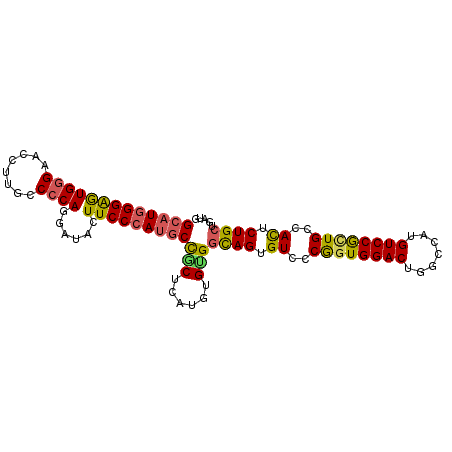
>2L_DroMel_CAF1 8483709 98 - 22407834 GCAUGGGAAUGGGAAUUUUGCCCCAGGAUACUUCCCAUGCCGCUCAUAUGUGGCAGUGUCCCGGUGGACUGACCAUGUCCGCUGCCACUCUGCUGAUG (((((((((((((........))))......)))))))))(((......)))((((.((..((((((((.......))))))))..)).))))..... ( -41.50) >DroSec_CAF1 23861 98 - 1 GCAUGGGAGUGGGAACCUGGCCCCAGGAUAUUUCCCAUGCCGCUCAUGUGUGGCAGUGUCCCGGUGGACUGGCCAUGUCCGCUGCCACUCUGCUGAUG (((((((.(((((((((((....))))....)))))))....)))))))..(((((.((..((((((((.......))))))))..)).))))).... ( -46.00) >DroSim_CAF1 24357 98 - 1 GCAUGGGAGUGGGAACCUGGCCCCAGGAUAUUUCCCAUGCCACUCAUGUGUGGCAGUGUCCCGGUGGACUGGCCAUGUCCACUGCCACUCUGCUGAUG (((((((.(((((((((((....))))....)))))))....)))))))..(((((.((..((((((((.......))))))))..)).))))).... ( -45.30) >DroEre_CAF1 31243 98 - 1 GCAUGGGAGUGGGAACCUUGCCCCAGGAUACUUCCCAUGCCGCCCAUGUGCGGCAGUGUGCCGCUGGACUGGCCAUGUCCGCUGCCACUCUGCUGAUG (((((((.((((((((((......)))....)))))))....))))))).((((((.((((.((.((((.......)))))).).))).))))))... ( -44.40) >DroYak_CAF1 41475 98 - 1 GCAUGGGAGUGGGAACCUUGCCCCAGGAUACUUCCCAUGCCGCUCAUGUGCGGCAGUGUCCCAGUGGACUGGCCAUGUCCGCUGCCACUCUGCUGAUG (((((((((((((........))))......)))))))))(((....)))((((((.((..((((((((.......))))))))..)).))))))... ( -44.40) >DroAna_CAF1 24231 98 - 1 GGAUUGGGAUGAGAACCGUGUCCCAGGAUGUUUCCCAAGCUGCUCAUGUGGGGUAGCGUGCCAGUGGACGGGCCAUGUCCGUUGCCAUUCUGAUGAUG (((((((........))).))))(((((((........(((((((.....))))))).........(((((((...)))))))..)))))))...... ( -32.40) >consensus GCAUGGGAGUGGGAACCUUGCCCCAGGAUACUUCCCAUGCCGCUCAUGUGUGGCAGUGUCCCGGUGGACUGGCCAUGUCCGCUGCCACUCUGCUGAUG (((((((((((((........))))......)))))))))(((......)))((((.((..((((((((.......))))))))..)).))))..... (-35.35 = -35.00 + -0.35)

| Location | 8,483,737 – 8,483,829 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 70.82 |
| Mean single sequence MFE | -36.67 |
| Consensus MFE | -13.55 |
| Energy contribution | -14.33 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.37 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.778699 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
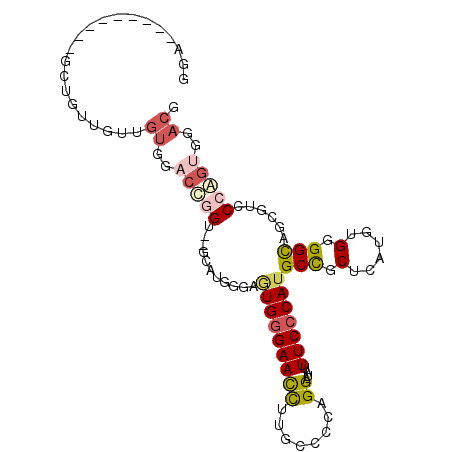
>2L_DroMel_CAF1 8483737 92 - 22407834 GUU---------GCUGUUGUUGUUGACCGGU--GCAUGGGAAUGGGAAUUUUGCCCCAGGAUACUUCCCAUGCCGCUCAUAUGUGGCAGUGUCCCGGUGGACU ...---------.........(((.(((((.--.((((((((((((........))))......))))).((((((......)))))))))..))))).))). ( -38.30) >DroPse_CAF1 27779 101 - 1 GGAGUGGGAAUGGGAGUUGGAGUG--CGAGUGCGAGUGCGAGUGGGAACCAUGGCCGAGGAUGUUUCCCAUGCUGCUCAUGUGGGGUAGCGUGCCAGUCGACG .((.(((.....((...(((..(.--((..(((....)))..)).)..)))...))..(((....)))((((((((((.....))))))))))))).)).... ( -33.60) >DroSec_CAF1 23889 92 - 1 GUU---------GCUGUUGUUGUUGACCGGU--GCAUGGGAGUGGGAACCUGGCCCCAGGAUAUUUCCCAUGCCGCUCAUGUGUGGCAGUGUCCCGGUGGACU ...---------.........(((.(((((.--.(((....(((((((((((....))))....)))))))(((((......))))).)))..))))).))). ( -41.10) >DroSim_CAF1 24385 92 - 1 GUU---------GCUGUUGUUGUUGACCGGU--GCAUGGGAGUGGGAACCUGGCCCCAGGAUAUUUCCCAUGCCACUCAUGUGUGGCAGUGUCCCGGUGGACU ...---------.........(((.(((((.--.(((....(((((((((((....))))....)))))))(((((......))))).)))..))))).))). ( -41.50) >DroAna_CAF1 24259 83 - 1 AGA----------CUG-------GGAGUGG---GGAUUGGGAUGAGAACCGUGUCCCAGGAUGUUUCCCAAGCUGCUCAUGUGGGGUAGCGUGCCAGUGGACG ..(----------(((-------(.(.(((---((((((((((.........))))))).....)))))).(((((((.....))))))).).)))))..... ( -31.90) >DroPer_CAF1 24677 101 - 1 GGAGUGGGAAUGGGAGUUGGAGUG--CGAGUGCGAGUGCGAGUGGGAACCAUGGCCGAGGAUGUUUCCCAUGCUGCUCAUGUGGGGUAGCGUGCCAGUCGACG .((.(((.....((...(((..(.--((..(((....)))..)).)..)))...))..(((....)))((((((((((.....))))))))))))).)).... ( -33.60) >consensus GGA_________GCUGUUGUUGUGGACCGGU__GCAUGGGAGUGGGAACCUUGCCCCAGGAUAUUUCCCAUGCCGCUCAUGUGGGGCAGCGUCCCAGUGGACG .....................((..(((((...........(((((((((........))....)))))))(((.(......).)))......)))))..)). (-13.55 = -14.33 + 0.78)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:40:22 2006