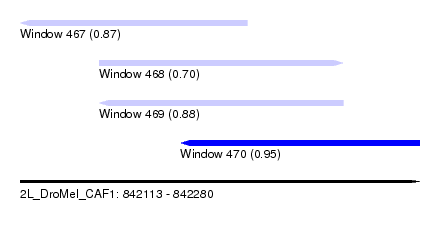
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 842,113 – 842,280 |
| Length | 167 |
| Max. P | 0.947414 |

| Location | 842,113 – 842,208 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 85.91 |
| Mean single sequence MFE | -24.86 |
| Consensus MFE | -18.06 |
| Energy contribution | -17.62 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.866757 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 842113 95 - 22407834 AAAUUAGCUCCGUGGAGUGGUCAUAACUAACUGUAAUAGCUGCUCCACUGGCCCCAGAGUCAUUCUUCUGCAUCUCCCCGA--GAUCAUCACCACAC ......(((..((((((..(..((.((.....)).))..)..)))))).)))..(((((......))))).(((((...))--)))........... ( -24.00) >DroSec_CAF1 7473 97 - 1 AAAUUAGCUGUGUGGAGUGGUCAUAACUAACUGUAAUAGCUGCUCCACUGGCCCCAGAGUCAUUCUUCUGCAUCUCCCCGAGAGAUCAUCUCCACAC .........(((((((((((((.........((((..((..((((...((....))))))....))..))))((((...))))))))).)))))))) ( -26.40) >DroSim_CAF1 8148 97 - 1 AAAUUAGCUGUGUGGAGUGGUCAUAACUAACUGUAAUAGCUGCUCCACUGGCCCCAGAGUCAUUCUGCUGCAUCUCCCCGAGAGAUCAUCUCCACAC .........(((((((((((((................((.((.....((((......))))....)).)).((((...))))))))).)))))))) ( -28.40) >DroEre_CAF1 7086 92 - 1 AAAUUAGCUCUUUGGAGUGGUCAUAACUAACUGUAAUAACUGCUCCACUGGCCCCGGAGCCAUUCCUCUCCAGCUCUGCGAGAUCU-----CUCCUC ......((.(..(((((..((.((.((.....)).)).))..)))))..)))..((((((............)))))).(((....-----)))... ( -24.30) >DroYak_CAF1 6299 92 - 1 AAAUUAGCUCUUUGGAGUGGUCAUAACUAACUGUAAUAACUGCUCCAGUGGCCCCAGAGUCAUUCUUCUCCAUCUCGUCGAGAUCU-----CUCCUC ......(((..((((((..((.((.((.....)).)).))..)))))).)))...((((....((((..(......)..)))).))-----)).... ( -21.20) >consensus AAAUUAGCUCUGUGGAGUGGUCAUAACUAACUGUAAUAGCUGCUCCACUGGCCCCAGAGUCAUUCUUCUGCAUCUCCCCGAGAGAUCAUC_CCACAC ...........((((((..((.((.((.....)).)).))..))))))((((......))))................................... (-18.06 = -17.62 + -0.44)

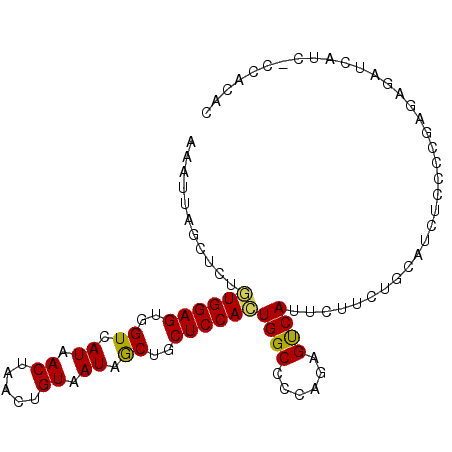
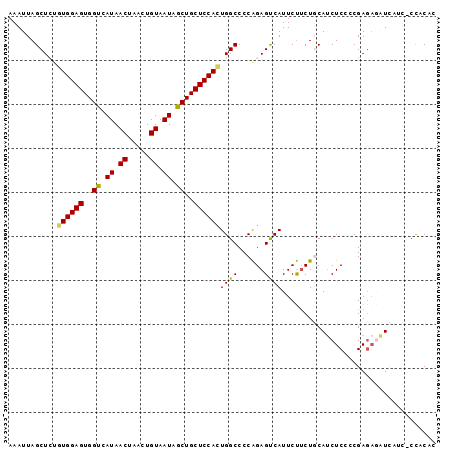
| Location | 842,146 – 842,248 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.21 |
| Mean single sequence MFE | -26.87 |
| Consensus MFE | -13.30 |
| Energy contribution | -13.44 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.699327 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 842146 102 + 22407834 -----UGACUCUGGGGCCAGUGGAGCAGCUAUUACAGUU-------------AGUUAUGACCACUCCACGGAGCUAAUUUGAUUUUUAGAGCCUCACUCUUUAGGCCAACAAACACUUUC -----.........((((.((((((..(.(((.((....-------------.)).))).)..))))))((..((((........))))..))..........))))............. ( -25.20) >DroSec_CAF1 7508 102 + 1 -----UGACUCUGGGGCCAGUGGAGCAGCUAUUACAGUU-------------AGUUAUGACCACUCCACACAGCUAAUUUAAUUUUUAGAGCCCCACUCUUUGGGCCAACAAACACUUUC -----......((((((..((((((..(.(((.((....-------------.)).))).)..))))))....((((........)))).))))))...((((......))))....... ( -24.30) >DroSim_CAF1 8183 90 + 1 -----UGACUCUGGGGCCAGUGGAGCAGCUAUUACAGUU-------------AGUUAUGACCACUCCACACAGCUAAUUUAAUUUUUAGAGCCC------------CAACAAACACUUUC -----......((((((..((((((..(.(((.((....-------------.)).))).)..))))))....((((........)))).))))------------))............ ( -23.60) >DroEre_CAF1 7116 99 + 1 -----UGGCUCCGGGGCCAGUGGAGCAGUUAUUACAGUU-------------AGUUAUGACCACUCCAAAGAGCUAAUUUAAUUUUUAGAGCCCCCCGCUUUGAGCC---AAACUCUUUC -----(((((((((((....(((((..(((((.((....-------------.)).)))))..)))))..(..((((........))))..).)))))....)))))---)......... ( -31.80) >DroYak_CAF1 6329 99 + 1 -----UGACUCUGGGGCCACUGGAGCAGUUAUUACAGUU-------------AGUUAUGACCACUCCAAAGAGCUAAUUUAAUUUUUAGAGCCCCACUCUUUAGGCC---AAGCACUUUC -----......((((((...(((((..(((((.((....-------------.)).)))))..))))).....((((........)))).))))))...........---.......... ( -24.20) >DroAna_CAF1 5976 115 + 1 ACCACUGAGGCUGGGGCCAUUGGAACAGCCAUUACAGUUAUUCCAGCCAGCCAGUUAGGGCCACUCCU-AGUGUUAAUUUCAUU-UUAGUGCCCUACUCUCUCAACA---CAACACUUUC .....((((((((((((...(((((.(((.......))).))))))))..)))))((((((.(((...-((((.......))))-..)))))))))....))))...---.......... ( -32.10) >consensus _____UGACUCUGGGGCCAGUGGAGCAGCUAUUACAGUU_____________AGUUAUGACCACUCCACAGAGCUAAUUUAAUUUUUAGAGCCCCACUCUUUAGGCC___AAACACUUUC ............(((((...(((((..(.(((.((..................)).))).)..))))).....((((........)))).)))))......................... (-13.30 = -13.44 + 0.14)

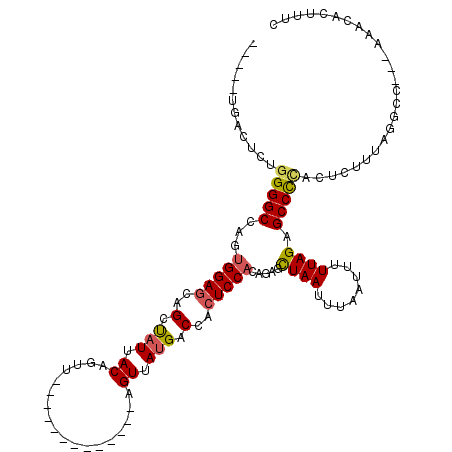
| Location | 842,146 – 842,248 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.21 |
| Mean single sequence MFE | -30.12 |
| Consensus MFE | -19.52 |
| Energy contribution | -20.35 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.877813 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 842146 102 - 22407834 GAAAGUGUUUGUUGGCCUAAAGAGUGAGGCUCUAAAAAUCAAAUUAGCUCCGUGGAGUGGUCAUAACU-------------AACUGUAAUAGCUGCUCCACUGGCCCCAGAGUCA----- .......((((..((((...((((.....))))..................((((((..(..((.((.-------------....)).))..)..)))))).)))).))))....----- ( -29.60) >DroSec_CAF1 7508 102 - 1 GAAAGUGUUUGUUGGCCCAAAGAGUGGGGCUCUAAAAAUUAAAUUAGCUGUGUGGAGUGGUCAUAACU-------------AACUGUAAUAGCUGCUCCACUGGCCCCAGAGUCA----- .....(.((((......)))).).(((((((((((........))))....((((((..(..((.((.-------------....)).))..)..)))))).)))))))......----- ( -29.80) >DroSim_CAF1 8183 90 - 1 GAAAGUGUUUGUUG------------GGGCUCUAAAAAUUAAAUUAGCUGUGUGGAGUGGUCAUAACU-------------AACUGUAAUAGCUGCUCCACUGGCCCCAGAGUCA----- ...........(((------------(((((((((........))))....((((((..(..((.((.-------------....)).))..)..)))))).)))))))).....----- ( -30.00) >DroEre_CAF1 7116 99 - 1 GAAAGAGUUU---GGCUCAAAGCGGGGGGCUCUAAAAAUUAAAUUAGCUCUUUGGAGUGGUCAUAACU-------------AACUGUAAUAACUGCUCCACUGGCCCCGGAGCCA----- .........(---(((((....(((((((((..............))))(..(((((..((.((.((.-------------....)).)).))..)))))..).)))))))))))----- ( -32.64) >DroYak_CAF1 6329 99 - 1 GAAAGUGCUU---GGCCUAAAGAGUGGGGCUCUAAAAAUUAAAUUAGCUCUUUGGAGUGGUCAUAACU-------------AACUGUAAUAACUGCUCCAGUGGCCCCAGAGUCA----- ......((((---........))))((((((((((........))))....((((((..((.((.((.-------------....)).)).))..)))))).)))))).......----- ( -27.00) >DroAna_CAF1 5976 115 - 1 GAAAGUGUUG---UGUUGAGAGAGUAGGGCACUAA-AAUGAAAUUAACACU-AGGAGUGGCCCUAACUGGCUGGCUGGAAUAACUGUAAUGGCUGUUCCAAUGGCCCCAGCCUCAGUGGU ...((((((.---((((....)).)).))))))..-...........((((-(((..(((........(((((..(((((((.((.....)).))))))).)))))))).))).)))).. ( -31.70) >consensus GAAAGUGUUU___GGCCCAAAGAGUGGGGCUCUAAAAAUUAAAUUAGCUCUGUGGAGUGGUCAUAACU_____________AACUGUAAUAGCUGCUCCACUGGCCCCAGAGUCA_____ .........................((((((((((........))))....((((((..((.((.((..................)).)).))..)))))).))))))............ (-19.52 = -20.35 + 0.83)

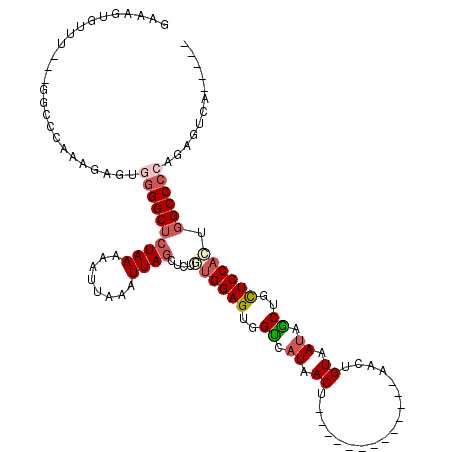
| Location | 842,180 – 842,280 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.09 |
| Mean single sequence MFE | -29.37 |
| Consensus MFE | -15.98 |
| Energy contribution | -16.48 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.947414 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 842180 100 - 22407834 --------CGAAACUGGACCACGCCACAUUGGCGUAGCUAGAAAGUGUUUGUUGGCCUAAAGAGUGAGGCUCUAAAAAUCAAAUUAGCUCCGUGGAGUGGUCAUAACU------------ --------........((((((.((((...((...((((((......(((...(((((........)))))...)))......)))))))))))).))))))......------------ ( -29.90) >DroSec_CAF1 7542 99 - 1 --------CGAAACUGGACCACGCCGCAUUG-UGCAGCUAGAAAGUGUUUGUUGGCCCAAAGAGUGGGGCUCUAAAAAUUAAAUUAGCUGUGUGGAGUGGUCAUAACU------------ --------........((((((.((((....-.((((((((.....((((...(((((........)))))....))))....)))))))))))).))))))......------------ ( -35.30) >DroSim_CAF1 8217 87 - 1 --------CGAAACUGGACCACGCCGCAUUG-UGCAGCUAGAAAGUGUUUGUUG------------GGGCUCUAAAAAUUAAAUUAGCUGUGUGGAGUGGUCAUAACU------------ --------........((((((.((((....-.((((((((.....((((.(((------------(....))))))))....)))))))))))).))))))......------------ ( -27.10) >DroEre_CAF1 7150 103 - 1 -UUCGUUAGAAAACUGGACCACGCCACAAUG-UGAAGCUAGAAAGAGUUU---GGCUCAAAGCGGGGGGCUCUAAAAAUUAAAUUAGCUCUUUGGAGUGGUCAUAACU------------ -...((((........((((((.((......-(((.((((((.....)))---))))))....((((((((..............)))))))))).)))))).)))).------------ ( -26.94) >DroYak_CAF1 6363 104 - 1 CAUCGUUGGGAAACUGGACCACGCCACAUUG-UGAAGCUAGAAAGUGCUU---GGCCUAAAGAGUGGGGCUCUAAAAAUUAAAUUAGCUCUUUGGAGUGGUCAUAACU------------ ....(((((....)).((((((........(-(.((((........))))---.))(((((((((.....................))))))))).))))))..))).------------ ( -29.30) >DroAna_CAF1 6016 105 - 1 --------GCAAACUGGUCCAGAUAG-UGAG-UGACCUUAGAAAGUGUUG---UGUUGAGAGAGUAGGGCACUAA-AAUGAAAUUAACACU-AGGAGUGGCCCUAACUGGCUGGCUGGAA --------.....(..(.((((.(((-(..(-(.(((((((..((((((.---((((....)).)).))))))..-..((.......))))-))).)).))....)))).)))))..).. ( -27.70) >consensus ________CGAAACUGGACCACGCCACAUUG_UGAAGCUAGAAAGUGUUU___GGCCCAAAGAGUGGGGCUCUAAAAAUUAAAUUAGCUCUGUGGAGUGGUCAUAACU____________ ................((((((.(((.........((((((............(((((........)))))............))))))...))).)))))).................. (-15.98 = -16.48 + 0.50)
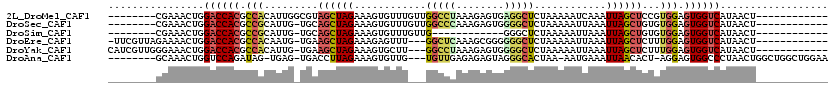
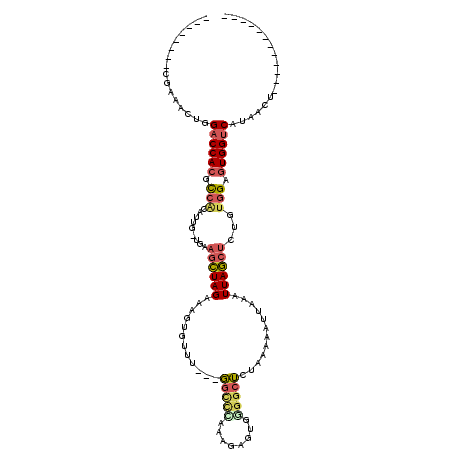
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:32:18 2006