| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,473,367 – 8,473,474 |
| Length | 107 |
| Max. P | 0.870783 |

| Location | 8,473,367 – 8,473,474 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 73.28 |
| Mean single sequence MFE | -35.39 |
| Consensus MFE | -11.94 |
| Energy contribution | -12.88 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.34 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.870783 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
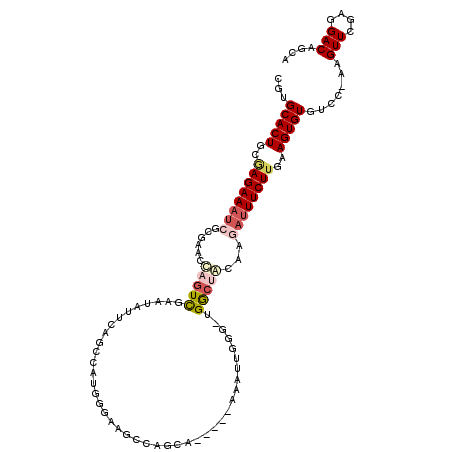
>2L_DroMel_CAF1 8473367 107 + 22407834 CGUGCACUGCGAGAAAUCGCGUACUAGUCGAAUAUUCAGCCAAGGGUAGGCAGCA-----AAAUUGGC-UGGCUACAAGAUUUCUUAAAGUGUGUCC-AAGUUCGAGGACAGCA .(((((((..((((((((..(((((((((((.......(((.......)))....-----...)))))-))).)))..))))))))..)))))((((-........)))).)). ( -36.54) >DroSec_CAF1 13878 107 + 1 CGUGCACUGCAAGAAAUCGCGAACUAGUUGAAUAUUCAGCCAUGGGAAGCCGGCC-----GAAUUGGG-UGGCUACAAGAUUUCUUGAAGUGUGUCC-AAGUUCGAGGACAGCA .(((((((.(((((((((.....((((((((....)))))..)))(.(((((.((-----.....)).-))))).)..))))))))).)))))((((-........)))).)). ( -38.70) >DroSim_CAF1 13908 107 + 1 CGUGCACUGCAAGAAAUCGCGAACUAGUUGAAUAUUCAGCCAUGGGAAGCCGGCA-----AAAUUGGG-UGACUACAAGAUUUCUUCAAGUGUGUCC-AAGUUCGAGGACAGCA .(((((((..((((((((((......(((((....)))))..(((....))))).-----....(((.-...)))...))))))))..)))))((((-........)))).)). ( -32.00) >DroEre_CAF1 14447 107 + 1 CAUGCACUGCGAGAAAUCGCGUAUCAGUCGCAUAUCCGGCCAUGCGCAGCCAGCC-----CAAUUGGC-UGGCCAACAGAUUUCUGGAAGUGUGUCC-AAGUUCGAGGACAGCA ...(((((.(.(((((((((((((..((((......)))).)))))).(((((((-----.....)))-)))).....))))))).).)))))((((-........)))).... ( -46.70) >DroYak_CAF1 31338 93 + 1 CGUGCACUGCGAGAAAUCGCGGUUCAGUCGAAUA--------------GCUAGCA-----UAAUUGAG-UGGCUAAAAAAUUUCUUAAAGUGUGGUG-AAGUUCGAGGACAGCA ..((((((((((....)))))))...(((...((--------------((((.((-----....))..-))))))...((((((............)-)))))....))).))) ( -25.30) >DroPer_CAF1 15164 108 + 1 CGUGCACUGAGAGAAACGGCAAAGCAAUU------CGGGCCACAACAAGGUAUCUCUCGUUAAUUGGAUUGGGGGUAAGCAUUCUUGGAGUGUGGCCUGGGUUCGAGGACAGCA .((.(.(((.......)))....(.((((------(((((((((.((((((((((((.(((.....))).))))))).....)))))...))))))))))))))..).)).... ( -33.10) >consensus CGUGCACUGCGAGAAAUCGCGAACCAGUCGAAUAUUCAGCCAUGGGAAGCCAGCA_____AAAUUGGG_UGGCUACAAGAUUUCUUGAAGUGUGUCC_AAGUUCGAGGACAGCA ...(((((..((((((((......(((((.........................................)))))...))))))))..))))).......(((....))).... (-11.94 = -12.88 + 0.94)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:40:17 2006