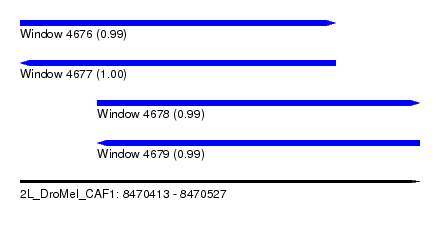
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,470,413 – 8,470,527 |
| Length | 114 |
| Max. P | 0.999922 |

| Location | 8,470,413 – 8,470,503 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 75.00 |
| Mean single sequence MFE | -21.75 |
| Consensus MFE | -10.24 |
| Energy contribution | -9.80 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.47 |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.986130 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8470413 90 + 22407834 AUCCACAUUCACAUCCACAUCCAUGGACAUAGCCAUGGCCAUAGCCGUAGCUCUUUGGCCAUGGCAAUUGCGAUUGCAUUUUCUUUAUCG .....((.((.((((((......))))....((((((((((.(((....)))...))))))))))...)).)).)).............. ( -28.70) >DroGri_CAF1 9885 69 + 1 AGCAGCA--AACAACCACA--CAUGGACA----------CAUAGAAAUAUCU-----GAGAUUGCAA--GCGAUUUGACUUUUCUUAUCG .((.(((--(.(..(((..--..))).((----------.(((....))).)-----).).))))..--))(((..((....))..))). ( -11.10) >DroSec_CAF1 11029 78 + 1 AUCCAC------AUCCACAUCCAUGGACA------UGGCCAUAGCCGUAGCUCUUUGGCCAUGGCAAUUGCGAUUGCAUUUUCUUUAUCG ......------.((((......))))((------((((((.(((....)))...))))))))(((((....)))))............. ( -23.60) >DroSim_CAF1 11129 78 + 1 AUCCAC------AUCCACAUCCAUGGACA------UGGCCAUAGCCGUAGCUCUUUGGCCAUGGCAAUUGCGAUUGCAUUUUCUUUAUCG ......------.((((......))))((------((((((.(((....)))...))))))))(((((....)))))............. ( -23.60) >consensus AUCCAC______AUCCACAUCCAUGGACA______UGGCCAUAGCCGUAGCUCUUUGGCCAUGGCAAUUGCGAUUGCAUUUUCUUUAUCG .............((((......))))................(((((.((......)).))))).....((((............)))) (-10.24 = -9.80 + -0.44)


| Location | 8,470,413 – 8,470,503 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 75.00 |
| Mean single sequence MFE | -27.30 |
| Consensus MFE | -16.42 |
| Energy contribution | -17.43 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.48 |
| Structure conservation index | 0.60 |
| SVM decision value | 4.57 |
| SVM RNA-class probability | 0.999922 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8470413 90 - 22407834 CGAUAAAGAAAAUGCAAUCGCAAUUGCCAUGGCCAAAGAGCUACGGCUAUGGCCAUGGCUAUGUCCAUGGAUGUGGAUGUGAAUGUGGAU .............(((.((((....((((((((((...(((....))).))))))))))...((((((....)))))))))).))).... ( -39.30) >DroGri_CAF1 9885 69 - 1 CGAUAAGAAAAGUCAAAUCGC--UUGCAAUCUC-----AGAUAUUUCUAUG----------UGUCCAUG--UGUGGUUGUU--UGCUGCU .(((..((....))..)))((--..((((((.(-----((((((......)----------))))....--)).)))))).--.)).... ( -13.50) >DroSec_CAF1 11029 78 - 1 CGAUAAAGAAAAUGCAAUCGCAAUUGCCAUGGCCAAAGAGCUACGGCUAUGGCCA------UGUCCAUGGAUGUGGAU------GUGGAU .................(((((.....((((((((...(((....))).))))))------))(((((....))))))------)))).. ( -28.20) >DroSim_CAF1 11129 78 - 1 CGAUAAAGAAAAUGCAAUCGCAAUUGCCAUGGCCAAAGAGCUACGGCUAUGGCCA------UGUCCAUGGAUGUGGAU------GUGGAU .................(((((.....((((((((...(((....))).))))))------))(((((....))))))------)))).. ( -28.20) >consensus CGAUAAAGAAAAUGCAAUCGCAAUUGCCAUGGCCAAAGAGCUACGGCUAUGGCCA______UGUCCAUGGAUGUGGAU______GUGGAU ((((............))))......(((((((((...(((....))).)))))........((((((....))))))......)))).. (-16.42 = -17.43 + 1.00)


| Location | 8,470,435 – 8,470,527 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 77.71 |
| Mean single sequence MFE | -24.68 |
| Consensus MFE | -15.49 |
| Energy contribution | -15.93 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.20 |
| Mean z-score | -3.14 |
| Structure conservation index | 0.63 |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.991103 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8470435 92 + 22407834 AUGGACA------UAGCCA------UGGCCAUAGCCGUAGCUCUUUGGCCAUGGCAAUUGCGAUUGCAUUUUCUUUAUCGCAUCCUGCACAAAUUACGAAAAUA ..(((..------..((((------((((((.(((....)))...))))))))))...((((((............)))))))))................... ( -31.90) >DroSec_CAF1 11045 86 + 1 AUGGACA------------------UGGCCAUAGCCGUAGCUCUUUGGCCAUGGCAAUUGCGAUUGCAUUUUCUUUAUCGCAUCCUGCACAAAUUACGAAAAUA .....((------------------((((((.(((....)))...))))))))(((..((((((............))))))...)))................ ( -26.40) >DroSim_CAF1 11145 86 + 1 AUGGACA------------------UGGCCAUAGCCGUAGCUCUUUGGCCAUGGCAAUUGCGAUUGCAUUUUCUUUAUCGCAUCCUGCACAAAUUACGAAAAUA .....((------------------((((((.(((....)))...))))))))(((..((((((............))))))...)))................ ( -26.40) >DroEre_CAF1 11448 104 + 1 ACAUCCAUGGCCAUAGCCAUAGCCAUUGCCAUUGCCGUAGCUCUUGGGCCAUGGCAAUUGCGAUUGCAUUUUCUUUAUCGCAUCCUGCACAAAUUACGAAAAUA ......(((((....))))).((.(((((.((((((((.(((....))).)))))))).))))).))((((((......((.....)).........)))))). ( -32.06) >DroAna_CAF1 11104 79 + 1 -------------------------AUGCUAUGGCCAUAGCUCUCUGGCUACGGCAAUUGCGAUUGCAUUUUCUUUAUCGCAUCCUGCACAAAUUACAAAAAUA -------------------------.(((..((((((........))))))..)))..((((((............))))))...................... ( -19.40) >DroPer_CAF1 12354 72 + 1 AUGGUCA------------------UGGCAAU------------UU--CCAUUGCGAUUGCUAUUGCAUUUUCUUUAUCGCAGCCUGCACAAAUUACAAAAAUA ..(((.(------------------(((....------------..--))))(((((((((....)))........)))))))))................... ( -11.90) >consensus AUGGACA__________________UGGCCAUAGCCGUAGCUCUUUGGCCAUGGCAAUUGCGAUUGCAUUUUCUUUAUCGCAUCCUGCACAAAUUACGAAAAUA ...........................(((((.((((........)))).)))))...((((((............))))))...................... (-15.49 = -15.93 + 0.44)

| Location | 8,470,435 – 8,470,527 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 77.71 |
| Mean single sequence MFE | -27.32 |
| Consensus MFE | -14.27 |
| Energy contribution | -14.10 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.99 |
| Structure conservation index | 0.52 |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.987580 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8470435 92 - 22407834 UAUUUUCGUAAUUUGUGCAGGAUGCGAUAAAGAAAAUGCAAUCGCAAUUGCCAUGGCCAAAGAGCUACGGCUAUGGCCA------UGGCUA------UGUCCAU .......(((.....))).(((((((((............)))))....((((((((((...(((....))).))))))------))))..------.)))).. ( -36.30) >DroSec_CAF1 11045 86 - 1 UAUUUUCGUAAUUUGUGCAGGAUGCGAUAAAGAAAAUGCAAUCGCAAUUGCCAUGGCCAAAGAGCUACGGCUAUGGCCA------------------UGUCCAU ................((((..((((((............)))))).))))((((((((...(((....))).))))))------------------))..... ( -29.10) >DroSim_CAF1 11145 86 - 1 UAUUUUCGUAAUUUGUGCAGGAUGCGAUAAAGAAAAUGCAAUCGCAAUUGCCAUGGCCAAAGAGCUACGGCUAUGGCCA------------------UGUCCAU ................((((..((((((............)))))).))))((((((((...(((....))).))))))------------------))..... ( -29.10) >DroEre_CAF1 11448 104 - 1 UAUUUUCGUAAUUUGUGCAGGAUGCGAUAAAGAAAAUGCAAUCGCAAUUGCCAUGGCCCAAGAGCUACGGCAAUGGCAAUGGCUAUGGCUAUGGCCAUGGAUGU ..........(((..((.....((((((............))))))...(((((((((....(((((..((....))..)))))..)))))))))))..))).. ( -34.30) >DroAna_CAF1 11104 79 - 1 UAUUUUUGUAAUUUGUGCAGGAUGCGAUAAAGAAAAUGCAAUCGCAAUUGCCGUAGCCAGAGAGCUAUGGCCAUAGCAU------------------------- ..............((((....((((((............))))))...((((((((......))))))))....))))------------------------- ( -22.40) >DroPer_CAF1 12354 72 - 1 UAUUUUUGUAAUUUGUGCAGGCUGCGAUAAAGAAAAUGCAAUAGCAAUCGCAAUGG--AA------------AUUGCCA------------------UGACCAU ......((((.....))))((.((((((........(((....)))))))))((((--..------------....)))------------------)..)).. ( -12.70) >consensus UAUUUUCGUAAUUUGUGCAGGAUGCGAUAAAGAAAAUGCAAUCGCAAUUGCCAUGGCCAAAGAGCUACGGCUAUGGCCA__________________UGUCCAU .((((((........(((.....))).....))))))((....))....(((((.(((..........))).)))))........................... (-14.27 = -14.10 + -0.17)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:40:16 2006