
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,443,836 – 8,443,979 |
| Length | 143 |
| Max. P | 0.645161 |

| Location | 8,443,836 – 8,443,956 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.11 |
| Mean single sequence MFE | -26.62 |
| Consensus MFE | -19.13 |
| Energy contribution | -19.80 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.643229 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
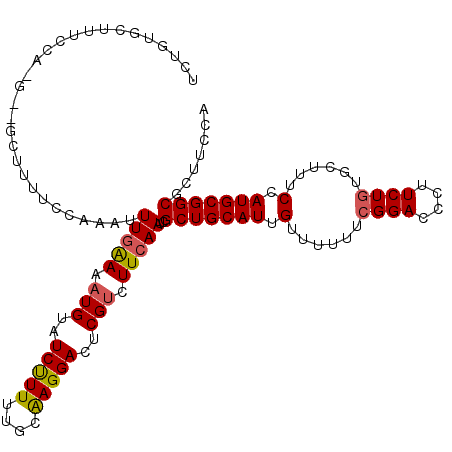
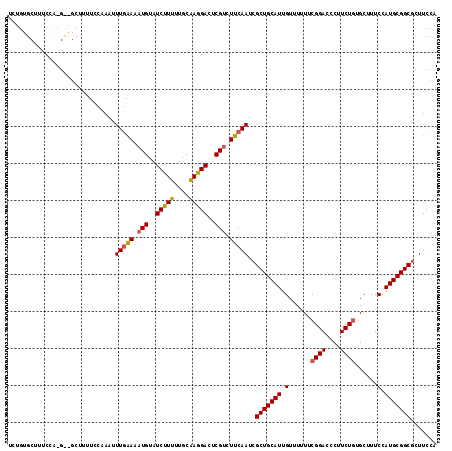
>2L_DroMel_CAF1 8443836 120 - 22407834 UGUAUCCUUUUGCGAGGACUCGUCUUCAAUCGCUGCAUUGUUUUUUCGGAAUCUUCUAUGCUUUCUAUGCGGCGUUUCCAAAUUUUGAAAAGUAUUCUUUUGCAAGGACUCGUCUUCAAU .(((......)))((((((..((((.....((((((((.........(((....))).........))))))))..........((((((((....))))).)))))))..))))))... ( -27.97) >DroSec_CAF1 3254 117 - 1 UGUAUCUUUUUCCAAGGAUACGCCUUCAAUCGCUGCAUUGUUUUUUCGGACCCUUCUGUGCUUUCCAUGCGGCUCUUCCAAAUUUGGAAAAGUAUUCUUUUCU---UUCUCGUCUUCAAU .((((((((....))))))))..........(((((((.(......((((....))))......).)))))))............(((((((....)))))))---.............. ( -24.90) >DroSim_CAF1 2806 117 - 1 UGUAUCUUUUUGCAAGGACUCGUCUUCAAUCGCUGCAUUGUUUUUUCGGACCCUUCUGUGCUUUCCAUGCGGCGCUUCCAAGUUUGGAAGAGUACUCUUUUCU---UUCUCGUCUUCAAU ((((......))))(((((...........((((((((.(......((((....))))......).))))))))((((((....)))))).............---.....))))).... ( -27.00) >consensus UGUAUCUUUUUGCAAGGACUCGUCUUCAAUCGCUGCAUUGUUUUUUCGGACCCUUCUGUGCUUUCCAUGCGGCGCUUCCAAAUUUGGAAAAGUAUUCUUUUCU___UUCUCGUCUUCAAU ((((......))))(((((............(((((((.(......((((....))))......).)))))))............(((((((....)))))))........))))).... (-19.13 = -19.80 + 0.67)



| Location | 8,443,876 – 8,443,979 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 81.32 |
| Mean single sequence MFE | -23.69 |
| Consensus MFE | -18.19 |
| Energy contribution | -18.53 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.645161 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8443876 103 - 22407834 -------UUUCCA------UUUUCCAAAUUUGAAAAUGUAUCCUUUUGCGAGGACUCGUCUUCAAUCGCUGCAUUGUUUUUUCGGAAUCUUCUAUGCUUUCUAUGCGGCGUUUCCA -------......------..........(((((.(((..(((((....)))))..))).))))).((((((((.........(((....))).........))))))))...... ( -22.37) >DroSec_CAF1 3291 116 - 1 UCUGUGCUUUCCAAGUCGCAUUUUCAUAGUUGGAAAUGUAUCUUUUUCCAAGGAUACGCCUUCAAUCGCUGCAUUGUUUUUUCGGACCCUUCUGUGCUUUCCAUGCGGCUCUUCCA ..((((((.....)).))))........((((((..(((((((((....)))))))))..)))))).(((((((.(......((((....))))......).)))))))....... ( -26.90) >DroSim_CAF1 2843 116 - 1 UCUGUGCUUUCCAUGCGGCUUUUCCAAAUUUAAAAAUGUAUCUUUUUGCAAGGACUCGUCUUCAAUCGCUGCAUUGUUUUUUCGGACCCUUCUGUGCUUUCCAUGCGGCGCUUCCA ((((......(.(((((((.................((((......))))((((....)))).....))))))).)......)))).......(((((........)))))..... ( -21.80) >consensus UCUGUGCUUUCCA_G__GCUUUUCCAAAUUUGAAAAUGUAUCUUUUUGCAAGGACUCGUCUUCAAUCGCUGCAUUGUUUUUUCGGACCCUUCUGUGCUUUCCAUGCGGCGCUUCCA .............................(((((.(((..(((((....)))))..))).)))))..(((((((.(......((((....))))......).)))))))....... (-18.19 = -18.53 + 0.34)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:40:05 2006