| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,434,304 – 8,434,410 |
| Length | 106 |
| Max. P | 0.752008 |

| Location | 8,434,304 – 8,434,410 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 77.36 |
| Mean single sequence MFE | -41.38 |
| Consensus MFE | -23.59 |
| Energy contribution | -24.48 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.34 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.708544 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
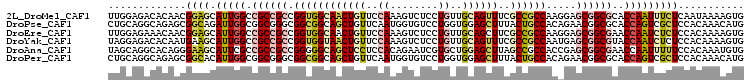
>2L_DroMel_CAF1 8434304 106 + 22407834 UUGGAGACACAACGGAGCAUUGGCCGCCGCCGGUGGCAACUGUUCCAAAGUCUCCUGUUGCAGUUUCGCCGCCAAGGAGCGGCGCACCAAUUUCUCAAUAAAAGUG ..((((((.....((((((...((((((...))))))...))))))...))))))(((((.((...((((((......)))))).........)))))))...... ( -39.80) >DroPse_CAF1 7095 106 + 1 CUGCAGGCAGAGCGGCAGAUUGGCGGCGGGCGGCGGCAGCUGUUCAAUGGUGUCCUGGUGGAGCUUUACUGCCACAGAACGGCGCACCAGUCGCUCCACAAACAUG ((((..((...)).))))...(((((((((((((....)))))))..((((((.(((.((..((......))..))...))).))))))))))))........... ( -40.30) >DroEre_CAF1 7284 106 + 1 UUGGAGAAACAACGGAGCAUUGGCCGCCGCCGGUGGCAACUGUUCCAAAGUCUCCUGUUGCAGCUUCGCCGCCAAGGAGCGGCGAACCAAUCUCUCCACAAAAGUG .((((((..((((((((((...((((((...))))))...))))))..((....))))))..(.((((((((......)))))))).)....))))))........ ( -43.60) >DroYak_CAF1 7368 106 + 1 UAGGAGACACAAUGAAGCAUUGGCCGCCGCCGGUGGUAACUGUUCCAAAGUCUCCUGUUGCAGUUUCGCCGCCAAUGAGCGGCGUACCAAUCUCUCCACAAAAGUG ..(((((..(((((...)))))(((((((..((((((((((((..((........))..))))))..))))))..)).))))).........)))))......... ( -38.90) >DroAna_CAF1 7700 106 + 1 UAGCAGGCACAGGGAAGCAUUCGCCGCCGCCGGGGGCAGCUCCUCCACAGAAUCGUGCUGGAGCUUAGCCGCCACCGAGCGGCGAACCAAUUUUUCCACAAAUGUG .......((((.(((((..((((((((((..((.(((((((((..(((......)))..))))))..))).))..)).))))))))......))))).....)))) ( -46.00) >DroPer_CAF1 7061 106 + 1 CUGCAGGCAGAGCGGCACAUUGGCGGCGGGCGGCGGCAGCUGUUCAAUGGUGUCCUGGUGGAGCUUUACUGCCACAGAACGGCGCACCAGUCGCUCCACAAACAUG ...((((((..((.((......)).))(((((((....))))))).....)).))))(((((((....(((...)))...(((......))))))))))....... ( -39.70) >consensus UUGCAGACACAACGGAGCAUUGGCCGCCGCCGGUGGCAACUGUUCCAAAGUCUCCUGUUGCAGCUUCGCCGCCAAAGAGCGGCGCACCAAUCUCUCCACAAAAGUG .............((((.(((((.((((((.((((((((((((..((........))..))))))..)))))).....))))))..)))))))))........... (-23.59 = -24.48 + 0.89)

| Location | 8,434,304 – 8,434,410 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 77.36 |
| Mean single sequence MFE | -42.58 |
| Consensus MFE | -23.76 |
| Energy contribution | -24.68 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.752008 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8434304 106 - 22407834 CACUUUUAUUGAGAAAUUGGUGCGCCGCUCCUUGGCGGCGAAACUGCAACAGGAGACUUUGGAACAGUUGCCACCGGCGGCGGCCAAUGCUCCGUUGUGUCUCCAA ..........((((....(((.(((((((....)))))))..)))(((((.((((...((((....((((((...))))))..))))..))))))))).))))... ( -41.40) >DroPse_CAF1 7095 106 - 1 CAUGUUUGUGGAGCGACUGGUGCGCCGUUCUGUGGCAGUAAAGCUCCACCAGGACACCAUUGAACAGCUGCCGCCGCCCGCCGCCAAUCUGCCGCUCUGCCUGCAG ..(((..(..(((((.(.((((((.((..(.((((((((.....(((....)))((....))....)))))))).)..)).)))..))).).)))))..)..))). ( -38.10) >DroEre_CAF1 7284 106 - 1 CACUUUUGUGGAGAGAUUGGUUCGCCGCUCCUUGGCGGCGAAGCUGCAACAGGAGACUUUGGAACAGUUGCCACCGGCGGCGGCCAAUGCUCCGUUGUUUCUCCAA ........(((((((...(.(((((((((....))))))))).).(((((.((((...((((....((((((...))))))..))))..)))))))))))))))). ( -49.80) >DroYak_CAF1 7368 106 - 1 CACUUUUGUGGAGAGAUUGGUACGCCGCUCAUUGGCGGCGAAACUGCAACAGGAGACUUUGGAACAGUUACCACCGGCGGCGGCCAAUGCUUCAUUGUGUCUCCUA (((....((((((..((((((.(((((((....((.((...((((((((.((....)))))...))))).)).))))))))))))))).)))))).)))....... ( -39.30) >DroAna_CAF1 7700 106 - 1 CACAUUUGUGGAAAAAUUGGUUCGCCGCUCGGUGGCGGCUAAGCUCCAGCACGAUUCUGUGGAGGAGCUGCCCCCGGCGGCGGCGAAUGCUUCCCUGUGCCUGCUA ((((...(.((((..(((.((.((((((.(((.(((((((...(((((.((......))))))).))))))).))))))))))).)))..)))))))))....... ( -49.00) >DroPer_CAF1 7061 106 - 1 CAUGUUUGUGGAGCGACUGGUGCGCCGUUCUGUGGCAGUAAAGCUCCACCAGGACACCAUUGAACAGCUGCCGCCGCCCGCCGCCAAUGUGCCGCUCUGCCUGCAG ..(((..(..((((((((((((.((.(..(.((((((((.....(((....)))((....))....)))))))).)..))))))))..))..)))))..)..))). ( -37.90) >consensus CACUUUUGUGGAGAGAUUGGUGCGCCGCUCCGUGGCGGCGAAGCUCCAACAGGAGACUUUGGAACAGCUGCCACCGGCGGCGGCCAAUGCUCCGCUGUGCCUCCAA .......((((((...(((((.(((((((...(((((((...........................)))))))..))))))))))))..))))))........... (-23.76 = -24.68 + 0.92)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:40:01 2006