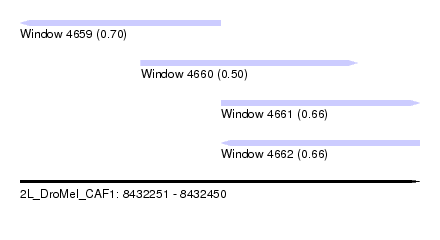
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,432,251 – 8,432,450 |
| Length | 199 |
| Max. P | 0.696694 |

| Location | 8,432,251 – 8,432,351 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 75.02 |
| Mean single sequence MFE | -38.18 |
| Consensus MFE | -19.09 |
| Energy contribution | -19.17 |
| Covariance contribution | 0.07 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.696694 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8432251 100 - 22407834 CUGCAGCGCAAGCAACUGGAGGCCUCGGCCCAGGAGGAGGC----------AGACCAG--CUGCAUCCCGAUACGGAUCCCA-----AUCCGGACUCGGGCGGCCAACUGCCAAACG .((((((....))..((((..(((((..(....)..)))))----------...))))--.)))).(((((..(((((....-----)))))...))))).(((.....)))..... ( -42.10) >DroSec_CAF1 5333 100 - 1 CUGCAGCGCAAGCAACUGGAGGCUUCGGCCCAGGAGGAGGC----------AGAUCAA--CUGCAUCCCGAUACGGAUCCCA-----AUCCGGACUCCGGCGACCAACUGCCAAACG .....((....))..((((..((....))))))..((((((----------((.....--)))).........(((((....-----)))))..))))((((......))))..... ( -36.50) >DroSim_CAF1 5315 100 - 1 CUGCAGCGCAAGAAACUGGAGGCUUCGGCCCAGGAGGAGGC----------AGACCAA--CUGCAUCCCGAUACGGAUCCCA-----AUCCGGACUCGGGCGACCAACUGCCAAACG ..(((((....)...((((..((....))))))..((..((----------((.....--))))..(((((..(((((....-----)))))...)))))...))..))))...... ( -35.10) >DroEre_CAF1 5321 100 - 1 CUGCAGCGCAAGCAACUGGAGGCUUCCGCUCAGGAGGAGGC----------AGACCAG--CUGCAACCUGAUACGGAUACCA-----GUCCAGACGCUGGCAGCCAACUACCAAACG (((((((((..(((.((((..((((((........))))))----------...))))--.)))..........((((....-----)))).).)))).)))).............. ( -36.10) >DroYak_CAF1 5454 100 - 1 CUGCAGCGUAAGCAACUGGAGGCUUCCGCUCAGGAGGAGGC----------AGACCAG--CUGCAUCCCGAUACGGAUCCCA-----AUCCAGACACUGGCGACCAACUGCCAAGCA ..((.((....))..(((((((..((((.((.(((....((----------((.....--)))).))).))..))))..)).-----.)))))....(((((......))))).)). ( -36.30) >DroAna_CAF1 5856 110 - 1 CAACAG------CUGCUGGAGGCAGCAGCCCAGGAGGCUGCGUUAGAAGCCAACCCAGAUCUGGAGCUAGGCAUAGAACCCAUCAGUGCCCAGUCGGUGG-AGACGUCCACCGAAUC ...(((------...((((.(((.((((((.....)))))).......)))...))))..)))...((.(((((.((.....)).))))).))(((((((-(....))))))))... ( -43.00) >consensus CUGCAGCGCAAGCAACUGGAGGCUUCGGCCCAGGAGGAGGC__________AGACCAG__CUGCAUCCCGAUACGGAUCCCA_____AUCCAGACUCUGGCGACCAACUGCCAAACG .....((....))..(((((.(((((..(....)..))))).......................((((......))))..........))))).....((((......))))..... (-19.09 = -19.17 + 0.07)

| Location | 8,432,311 – 8,432,419 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 86.36 |
| Mean single sequence MFE | -40.20 |
| Consensus MFE | -31.15 |
| Energy contribution | -32.54 |
| Covariance contribution | 1.39 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.77 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8432311 108 + 22407834 CCUCCUCCUGGGCCGAGGCCUCCAGUUGCUUGCGCUGCAGCUCCAGGCGAUUGAGCAACUCAACCGCGGUCUCCUGGAUCUGCUGCUGCAGCUCGUGCGCAUGCUCGU .......((((((....))..))))..((.(((((.(((((((((((.(((((.............))))).))))))...))))).)).((....))))).)).... ( -40.12) >DroSec_CAF1 5393 108 + 1 CCUCCUCCUGGGCCGAAGCCUCCAGUUGCUUGCGCUGCAGCUCCAGGCGAUUGAGCAACUCAACCGCAGUCUCCUGGAUCUGCUGCUGCAGCUCGUGCGCAUGCUCAU ........(((((....((..(.((((((....((.((((.((((((.(((((.............))))).)))))).)))).)).)))))).).))....))))). ( -41.02) >DroSim_CAF1 5375 108 + 1 CCUCCUCCUGGGCCGAAGCCUCCAGUUUCUUGCGCUGCAGCUCCAGGCGAUUGAGCAACUCAACCGCAGUCUCCUGGAUCUGCUGCUGCAGCUCGUGCGCAUGCUCAU ........(((((.(((((.....))))).(((((.(((((((((((.(((((.............))))).))))))...))))).)).((....))))).))))). ( -40.22) >DroEre_CAF1 5381 108 + 1 CCUCCUCCUGAGCGGAAGCCUCCAGUUGCUUGCGCUGCAGCUCCAGGCGAUUGAGCAACUCAACCGCAGUCUCCUGGAUCUGCUGCUGCAGCUCGUGCGCAUGCUCGU .........(((((...((..(.((((((....((.((((.((((((.(((((.............))))).)))))).)))).)).)))))).).))...))))).. ( -43.42) >DroWil_CAF1 6375 102 + 1 CUUCCU---GGAGUUGUUCGUCGA---GCUUUUGCUGCAACUCCAUUUGAUUGACCAACUGGACCACAUGCUCCUCGAUCUGCUGUUGCAAUUGAUGGGCGUGCUCAU ..((((---((((((((......(---((....)))))))))))).(((......)))..)))...(((((((.(((((.(((....)))))))).)))))))..... ( -33.00) >DroYak_CAF1 5514 108 + 1 CCUCCUCCUGAGCGGAAGCCUCCAGUUGCUUACGCUGCAGCUCCAGGCGAUUGAGCAACUCAACCGCAGUCUCCUGGAUCUGCUGCUGCAGCUCGUGCGCAUGCUCGU .........(((((...((..(.((((((....((.((((.((((((.(((((.............))))).)))))).)))).)).)))))).).))...))))).. ( -43.42) >consensus CCUCCUCCUGGGCCGAAGCCUCCAGUUGCUUGCGCUGCAGCUCCAGGCGAUUGAGCAACUCAACCGCAGUCUCCUGGAUCUGCUGCUGCAGCUCGUGCGCAUGCUCAU .........((((....((..(.((((((....((.((((.((((((.(((((.............))))).)))))).)))).)).)))))).).))....)))).. (-31.15 = -32.54 + 1.39)


| Location | 8,432,351 – 8,432,450 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 81.92 |
| Mean single sequence MFE | -39.04 |
| Consensus MFE | -17.72 |
| Energy contribution | -17.61 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.13 |
| Mean z-score | -3.03 |
| Structure conservation index | 0.45 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.657724 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8432351 99 + 22407834 CUCCAGGCGAUUGAGCAACUCAACCGCGGUCUCCUGGAUCUGCUGCUGCAGCUCGUGCGCAUGCUCGUGCAUGUGGCU---------GUGCUGAUGCUGCACAAUUUU .((((((.(((((.............))))).))))))..(((.((..((((.((.((((((((....)))))).)))---------).))))..)).)))....... ( -37.02) >DroPse_CAF1 5617 108 + 1 UUCCAUCUGGUUGACCAGCUGGACCACAUGCUCCUCGAUCUGCUGCUGCAGCUCGUGAGCAUGCUCGUGCAUAUGGUUGUGAAAAUUGUGCUGAUGCUGCACAAUUUU .((((.((((....)))).)))).((((.((((..(((.((((....)))).))).))))((((....)))).....))))((((((((((.......)))))))))) ( -44.30) >DroSec_CAF1 5433 99 + 1 CUCCAGGCGAUUGAGCAACUCAACCGCAGUCUCCUGGAUCUGCUGCUGCAGCUCGUGCGCAUGCUCAUGCAUGUGACU---------GUGCUGAUGCUGCACAAUUUU .((((((.(((((.............))))).))))))..(((.((..((((.((..(((((((....)))))))..)---------).))))..)).)))....... ( -38.52) >DroSim_CAF1 5415 99 + 1 CUCCAGGCGAUUGAGCAACUCAACCGCAGUCUCCUGGAUCUGCUGCUGCAGCUCGUGCGCAUGCUCAUGCAUGUGGCU---------GUGCUGAUGCUGCACAAUUUU .((((((.(((((.............))))).)))))).....((.(((((((((.(((((.((.(((....))))))---------))))))).))))))))..... ( -38.02) >DroWil_CAF1 6409 108 + 1 CUCCAUUUGAUUGACCAACUGGACCACAUGCUCCUCGAUCUGCUGUUGCAAUUGAUGGGCGUGCUCAUGCAGAUGAUUGUGAAAAUUAUGCUGAUGCUGCACAAUUUU .((((.(((......))).))))......((((.(((((.(((....)))))))).))))((((.(((.(((((((((.....)))))).))))))..))))...... ( -32.10) >DroPer_CAF1 5590 108 + 1 UUCCAUCUGGUUGACCAGCUGGACCACAUGCUCCUCGAUCUGCUGCUGCAGCUCGUGAGCAUGCUCGUGCAUAUGGUUGUGAAAAUUGUGCUGAUGCUGCACAAUUUU .((((.((((....)))).)))).((((.((((..(((.((((....)))).))).))))((((....)))).....))))((((((((((.......)))))))))) ( -44.30) >consensus CUCCAGGCGAUUGACCAACUCAACCACAGGCUCCUCGAUCUGCUGCUGCAGCUCGUGCGCAUGCUCAUGCAUAUGGCU_________GUGCUGAUGCUGCACAAUUUU ........(((((.......................((.((((....)))).))....(((.(((((.((((...............))))))).))))).))))).. (-17.72 = -17.61 + -0.11)


| Location | 8,432,351 – 8,432,450 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 81.92 |
| Mean single sequence MFE | -38.10 |
| Consensus MFE | -16.71 |
| Energy contribution | -15.22 |
| Covariance contribution | -1.50 |
| Combinations/Pair | 1.39 |
| Mean z-score | -3.12 |
| Structure conservation index | 0.44 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.655787 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8432351 99 - 22407834 AAAAUUGUGCAGCAUCAGCAC---------AGCCACAUGCACGAGCAUGCGCACGAGCUGCAGCAGCAGAUCCAGGAGACCGCGGUUGAGUUGCUCAAUCGCCUGGAG ....((((((.((....))..---------.....(((((....))))).)))))).((((....)))).((((((....)(((((((((...)))))))))))))). ( -36.60) >DroPse_CAF1 5617 108 - 1 AAAAUUGUGCAGCAUCAGCACAAUUUUCACAACCAUAUGCACGAGCAUGCUCACGAGCUGCAGCAGCAGAUCGAGGAGCAUGUGGUCCAGCUGGUCAACCAGAUGGAA ((((((((((.......)))))))))).....((((((((..(((....))).(((.((((....)))).)))....))))))))((((.((((....)))).)))). ( -45.60) >DroSec_CAF1 5433 99 - 1 AAAAUUGUGCAGCAUCAGCAC---------AGUCACAUGCAUGAGCAUGCGCACGAGCUGCAGCAGCAGAUCCAGGAGACUGCGGUUGAGUUGCUCAAUCGCCUGGAG ....((((((.((....))..---------.....(((((....))))).)))))).((((....)))).(((((....))(((((((((...)))))))))..))). ( -37.00) >DroSim_CAF1 5415 99 - 1 AAAAUUGUGCAGCAUCAGCAC---------AGCCACAUGCAUGAGCAUGCGCACGAGCUGCAGCAGCAGAUCCAGGAGACUGCGGUUGAGUUGCUCAAUCGCCUGGAG ....((((((.((....))..---------.....(((((....))))).)))))).((((....)))).(((((....))(((((((((...)))))))))..))). ( -37.00) >DroWil_CAF1 6409 108 - 1 AAAAUUGUGCAGCAUCAGCAUAAUUUUCACAAUCAUCUGCAUGAGCACGCCCAUCAAUUGCAACAGCAGAUCGAGGAGCAUGUGGUCCAGUUGGUCAAUCAAAUGGAG ((((((((((.......))))))))))((((......(((....))).((((.((..((((....))))...)))).)).)))).((((.((((....)))).)))). ( -26.80) >DroPer_CAF1 5590 108 - 1 AAAAUUGUGCAGCAUCAGCACAAUUUUCACAACCAUAUGCACGAGCAUGCUCACGAGCUGCAGCAGCAGAUCGAGGAGCAUGUGGUCCAGCUGGUCAACCAGAUGGAA ((((((((((.......)))))))))).....((((((((..(((....))).(((.((((....)))).)))....))))))))((((.((((....)))).)))). ( -45.60) >consensus AAAAUUGUGCAGCAUCAGCAC_________AACCACAUGCACGAGCAUGCGCACGAGCUGCAGCAGCAGAUCCAGGAGAAUGCGGUCCAGUUGCUCAAUCACAUGGAG ......((((.......))))...........((((((((....))))).((((..((((((....(........)....))))))...).))).........))).. (-16.71 = -15.22 + -1.50)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:39:59 2006