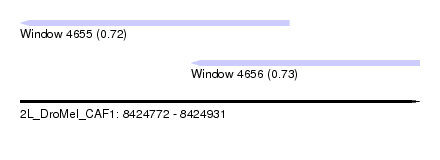
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,424,772 – 8,424,931 |
| Length | 159 |
| Max. P | 0.728471 |

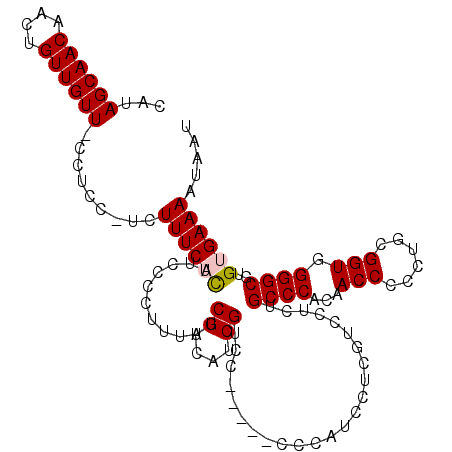
| Location | 8,424,772 – 8,424,879 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 88.00 |
| Mean single sequence MFE | -20.93 |
| Consensus MFE | -17.76 |
| Energy contribution | -17.92 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.718187 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8424772 107 - 22407834 CAUAGCAACAACUGUUGUU-CCUUCCUCUUUCACUUCCCCUUUAUCGUCAUCGUCC------CCAUCCUCGUCCUCUGCCCACAACCCCCUGCGGUGGGGCGUGUGAAAAUAAU ...((((((....))))))-.....................((((..((((((.((------((((((.......................).))))))))).))))..)))). ( -21.40) >DroSec_CAF1 22883 108 - 1 CAUAGCAACAACUGUUGUU-CCUCCCUCUUUCACUUCCCCUUUAUCGUCAUCGUCC-----CCCAUCCUCGUCCUCUGCCCACAACCCCCUGCGGUGGGGCGUGUGAAAAUAAU ...((((((....))))))-........((((((...........((....))...-----................((((...(((......))).))))..))))))..... ( -20.30) >DroSim_CAF1 22786 108 - 1 CAUAGCAACAACUGUUGUU-UCUCCUUCUUUCACUUCCCCUUUAUCGUCAUCGUCC-----CCCAUCCUCGUCCUCUGCCCACAACCCCAUGCGGUGGGGCGUGUGAAAAUAAU ...((((((....))))))-.....................((((..(((.((..(-----((((.((.(((....((....)).....))).)))))))..)))))..)))). ( -20.40) >DroEre_CAF1 23181 112 - 1 CCUAGCAACAACUGUUGUUCGCCC--CCUUUCUUUUCCCCUUUAUCGUCAUCGUCAUCGUCCCCAUCCUCGUCCUCUGCCCACAACCCCCUGUGGUGGGGCGUGUGAAAAUAAU ...((((((....)))))).....--...........................((((((.(((((.....((.....))(((((......)))))))))))).))))....... ( -21.80) >DroYak_CAF1 23321 111 - 1 CCUAGCAACAACUGUUGUU-ACUC--CCUUUCUUCUCUUCUUCUUCGUCAUCGUCCCCAUCUCCAUUCUCGUCCUCUGCCCACAACCCCCUGCGGUGGGGCGUGUGAAAAUAAU ..(((((((....))))))-)...--.................((((.((.((.(((((((................................)))))))))))))))...... ( -20.75) >consensus CAUAGCAACAACUGUUGUU_CCUCC_UCUUUCACUUCCCCUUUAUCGUCAUCGUCC_____CCCAUCCUCGUCCUCUGCCCACAACCCCCUGCGGUGGGGCGUGUGAAAAUAAU ...((((((....)))))).........((((((...........((....))........................((((...(((......))).))))..))))))..... (-17.76 = -17.92 + 0.16)


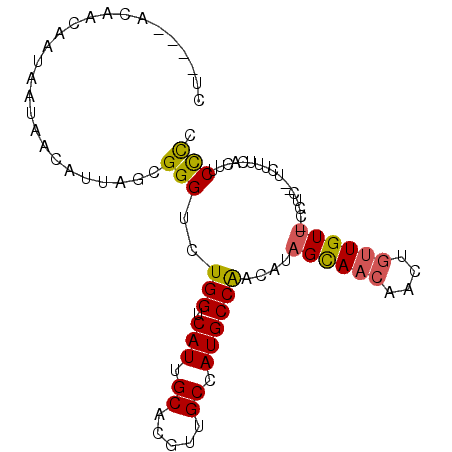
| Location | 8,424,840 – 8,424,931 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 78.68 |
| Mean single sequence MFE | -17.37 |
| Consensus MFE | -11.74 |
| Energy contribution | -11.52 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.728471 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8424840 91 - 22407834 CU----ACAACAAUAAUAACAUUAGCGGGUCUGGUCAUUGCACGUUGCUAUGCCAACAUAGCAACAACUGUUGUU-CCUUCCUCUUUCACUUCCCC ..----((((((............((((.........))))..(((((((((....)))))))))...)))))).-.................... ( -19.50) >DroSec_CAF1 22952 91 - 1 CU----ACAACAAUAAUAACAUUAGCGGGUCUGGUCAUUGCACGUUGCCAUGCCAACAUAGCAACAACUGUUGUU-CCUCCCUCUUUCACUUCCCC ..----.................((.(((..(((.(((.((.....)).))))))....((((((....))))))-...))).))........... ( -16.00) >DroSim_CAF1 22855 91 - 1 CU----ACAACAAUAAUAACAUUAGCGGGUCUGGUCAUUGCACGUUGCCAUGCCGACAUAGCAACAACUGUUGUU-UCUCCUUCUUUCACUUCCCC ..----((((((.((((...))))((..(((.((.(((.((.....)).))))))))...))......)))))).-.................... ( -15.80) >DroEre_CAF1 23255 90 - 1 CU----ACAACAAUAAUAACAUUAACGGGUCUGGUCAUUGCACGUUGCCAUGCCAACCUAGCAACAACUGUUGUUCGCCC--CCUUUCUUUUCCCC ..----....................((((.(((.(((.((.....)).))))))....((((((....)))))).))))--.............. ( -16.90) >DroYak_CAF1 23395 89 - 1 CU----ACAACAAUAAUAACAUUAACGGGUCUGGUCAUUGCACGUUGCCAUGCCAACCUAGCAACAACUGUUGUU-ACUC--CCUUUCUUCUCUUC ..----....................(((..(((.(((.((.....)).))))))...(((((((....))))))-)..)--))............ ( -17.20) >DroPer_CAF1 23080 77 - 1 CGUAACGCAACGGG--------GAUCGGGUCUGGUCAUCGCACGUUGCCAUGCCAACGAUGUA------GUUGUC-UCU----CUGCCGCUGCUUC ......(((.(((.--------(((((....)))))...((((((((......))))).))).------......-...----...))).)))... ( -18.80) >consensus CU____ACAACAAUAAUAACAUUAGCGGGUCUGGUCAUUGCACGUUGCCAUGCCAACAUAGCAACAACUGUUGUU_CCUC__UCUUUCACUUCCCC ..........................(((..(((.(((.((.....)).))))))....((((((....)))))).................))). (-11.74 = -11.52 + -0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:39:54 2006