
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,423,691 – 8,423,825 |
| Length | 134 |
| Max. P | 0.994456 |

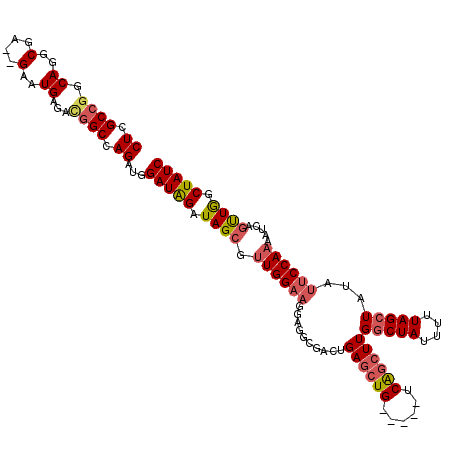
| Location | 8,423,691 – 8,423,802 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 88.46 |
| Mean single sequence MFE | -41.26 |
| Consensus MFE | -25.68 |
| Energy contribution | -26.60 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.542893 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8423691 111 + 22407834 CUCGCCGGCAGGCGA--GAAUGAGACGGCCAGAUGGAUAGAUAGCGUUGGAAGGAGGUGACUGAGCUG-----UCAGCUUGGCUAUUUUUAGCUAUAUUCCAAAAUCACUUGGCUAUC ((((((....)))))--).....((..(((((.(((...(....).((((((.(((.((((......)-----))).)))(((((....)))))...))))))..))).)))))..)) ( -38.90) >DroSec_CAF1 21888 116 + 1 CUCGCCGGCAGGCGA--GAAUGAGAUGGCCAGAUGGAUAGAUAGCGUUGGAAGGAGGCGACUGAGCUGUCAUGUCGGCUUGGCUAUUUUUAGCUAUAUUCCAAAAUCAGUUGGCUAUC ((((((....)))))--).....(((((((((...(((........((((((.(((.(((((((....))).)))).)))(((((....)))))...)))))).)))..))))))))) ( -45.40) >DroSim_CAF1 21726 118 + 1 CUCGCCGGCAGGCGAGAGAAUGAGAUGGCCAGAUGGAUAGAUAGCGUUGGAAGGAGGCGACUGAGCUGCCAUGUCAGCUUGGCUAUUUUUAGCUAUAUUCCAAAAUCAGUUGGCUAUC ((((((....)))))).......(((((((((.((((...(((((....(((((.(((...(((((((......)))))))))).))))).)))))..)))).......))))))))) ( -43.10) >DroEre_CAF1 22136 108 + 1 CUCGCGGGCAGGCGA--GAAUGAGACGGCCAGAUGGAUGGAUAGCGUUGG---GAUGCGGCUGAACUG-----UCAUCUUGACUAUUUUUAGCUAUAUUCCAAAAUCAGCUAUCUAUC (((((......))))--).................((((((((((.((((---((((..((((((..(-----((.....)))....))))))..)))))))).....)))))))))) ( -40.60) >DroYak_CAF1 22168 111 + 1 CUCGCCAGCAGGCGA--GAAUGAGACGGCCAGAUGGAUAGAUAGCGCUGGAAGGAGGCGACUGAGCUG-----UCAGCUUGGCUAUUUAUAGCUAAAUUCCAAUAUCAGUUAUCUAUC ((((((....)))))--).................(((((((((((((((.....(((......))).-----)))))(((((((....)))))))............)))))))))) ( -38.30) >consensus CUCGCCGGCAGGCGA__GAAUGAGACGGCCAGAUGGAUAGAUAGCGUUGGAAGGAGGCGACUGAGCUG_____UCAGCUUGGCUAUUUUUAGCUAUAUUCCAAAAUCAGUUGGCUAUC ((.((((.((..(....)..))...)))).))...(((((.((((.((((((..........((((((......))))))(((((....)))))...)))))).....)))).))))) (-25.68 = -26.60 + 0.92)

| Location | 8,423,727 – 8,423,825 |
|---|---|
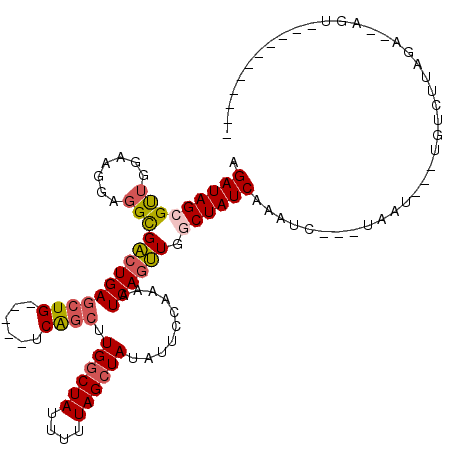
| Length | 98 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.88 |
| Mean single sequence MFE | -28.87 |
| Consensus MFE | -18.48 |
| Energy contribution | -19.08 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.64 |
| SVM decision value | 2.48 |
| SVM RNA-class probability | 0.994456 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8423727 98 + 22407834 AGAUAGCGUUGGAAGGAGGUGACUGAGCUG-----UCAGCUUGGCUAUUUUUAGCUAUAUUCCAAAAUCACUUGGCUAUCAAAUC---UAAU---UGUCUCAGAAUAGU----------- .((((((.((((((.(((.((((......)-----))).)))(((((....)))))...))))))...(....))))))).....---....---..............----------- ( -26.10) >DroSec_CAF1 21924 101 + 1 AGAUAGCGUUGGAAGGAGGCGACUGAGCUGUCAUGUCGGCUUGGCUAUUUUUAGCUAUAUUCCAAAAUCAGUUGGCUAUCAAAUC---UAAU---UGUCUUAGA--AGU----------- .((((((.((((((.(((.(((((((....))).)))).)))(((((....)))))...))))))...(....)))))))...((---(((.---....)))))--...----------- ( -30.30) >DroSim_CAF1 21764 101 + 1 AGAUAGCGUUGGAAGGAGGCGACUGAGCUGCCAUGUCAGCUUGGCUAUUUUUAGCUAUAUUCCAAAAUCAGUUGGCUAUCAAAUC---UAGU---UGUCUUAGA--AGU----------- (((((((.((((....((.(((((((((((......)))).((((((....))))))..........))))))).)).))))...---..))---)))))....--...----------- ( -29.50) >DroEre_CAF1 22172 108 + 1 GGAUAGCGUUGG---GAUGCGGCUGAACUG-----UCAUCUUGACUAUUUUUAGCUAUAUUCCAAAAUCAGCUAUCUAUCAAAUUCGCUGGUCAAUAUUUUAAA--AAUCGUUCUCCU-- (((.((((((((---((((..((((((..(-----((.....)))....))))))..)))))))).((((((..............))))))............--...)))).))).-- ( -30.84) >DroYak_CAF1 22204 110 + 1 AGAUAGCGCUGGAAGGAGGCGACUGAGCUG-----UCAGCUUGGCUAUUUAUAGCUAAAUUCCAAUAUCAGUUAUCUAUCAAAUU---UGAUAAGUCUCUUAAC--ACUACUUCUAUUAA .(((((...((..(((((((((((((((..-----...))(((((((....))))))).........))))))...(((((....---))))).)))))))..)--)......))))).. ( -27.60) >consensus AGAUAGCGUUGGAAGGAGGCGACUGAGCUG_____UCAGCUUGGCUAUUUUUAGCUAUAUUCCAAAAUCAGUUGGCUAUCAAAUC___UAAU___UGUCUUAGA__AGU___________ .(((((((((.......)))((((((((((......)))).((((((....))))))..........)))))).))))))........................................ (-18.48 = -19.08 + 0.60)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:39:52 2006