| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 840,612 – 840,722 |
| Length | 110 |
| Max. P | 0.864253 |

| Location | 840,612 – 840,722 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.21 |
| Mean single sequence MFE | -38.43 |
| Consensus MFE | -14.88 |
| Energy contribution | -14.80 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.39 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.864253 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
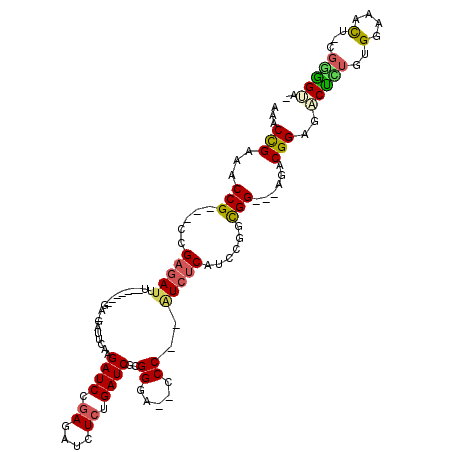
>2L_DroMel_CAF1 840612 110 - 22407834 AAACUGAAACCG---CCGAGAUUCGAGAUUCGA-GAUUCAAGAUCCGAGAUCUCUGAUCGCGGGU--CCCGGGUCUCAUCCGGCGG---AGACGGUGACUAUGUGGAAACUGUGAGGUG- ..((((...(((---(((.(((..((((((((.-(((((..((((.((....)).))))..))))--).)))))))))))))))))---...))))..((....(....)....))...- ( -42.30) >DroSec_CAF1 6014 100 - 1 AAACCGAAACCG---CCGAGAUUU-------GA-GAUUCAAGAUCCGAGAUCUCUGAUCGCGGGA--CCC--AUCUCAUCCGGCGG---AGACGGAGACUCUGAGGAAACU-CGGGGUG- ...(((...(((---(((.((..(-------((-(((....(.((((.((((...)))).)))).--)..--))))))))))))))---...)))..((((((((....))-)))))).- ( -45.20) >DroSim_CAF1 6691 100 - 1 AAACCGAAACCG---CCGAGAUUC-------GA-GAUUCAAGAUCCGAGAUCUCUGAUCGCGGGA--CCC--AUCUCAUCCGGCGG---AGACGGAGACUCUGUGGAAACU-CGGGGUG- ...(((...(((---(((.(((..-------((-(((....(.((((.((((...)))).)))).--)..--))))))))))))))---...)))..((((((.(....).-)))))).- ( -40.50) >DroEre_CAF1 5732 93 - 1 AAACCGAAACCG---CCGAGAUUU-------G--------UGAUCCGAGAUCUCUGAUCGUGGUG--CCC--AUCUCAUCCGCCGG---AGGCGGACGCUUUGUGGAUACU-CGGGGCA- ........((((---(.(((((((-------.--------.......))))))).....)))))(--(((--.....((((((..(---((((....)))))))))))...-..)))).- ( -31.70) >DroYak_CAF1 4834 93 - 1 AAACCGAAACCG---CCGAGAUUU-------C--------AGAUCCGAGAUCUCUGAUCGUGGUG--CCC--AUCUCAUCCGCCGG---AGACGGCGACGUUGUGGAAACU-CGGCGCA- ..........((---(((((.(((-------(--------(((((.((....)).))))((((..--.))--))......(((((.---...)))))......))))).))-)))))..- ( -36.00) >DroAna_CAF1 4674 116 - 1 AAACCGAAACCGAAACCGAAACCGAAACAGCGACGAUCCGAGAUCCAAGAUCUCAGAUCUGGGAAUCUCC--AUC-CAUCCGGUGGCUCGGACGGAGGCUGUGUGGAAAUU-CCCAGAAG ....((....((....))....((......)).)).((.((((((...)))))).))(((((((((.(((--(((-(.(((((....))))).))).......)))).)))-)))))).. ( -34.91) >consensus AAACCGAAACCG___CCGAGAUUU_______GA_GAUUCAAGAUCCGAGAUCUCUGAUCGCGGGA__CCC__AUCUCAUCCGGCGG___AGACGGAGACUCUGUGGAAACU_CGGGGUA_ ...(((...(((.....(((((...................((((.((....)).))))..((.....))..)))))......)))......)))..(((((..(....)...))))).. (-14.88 = -14.80 + -0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:32:12 2006