
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,421,632 – 8,421,742 |
| Length | 110 |
| Max. P | 0.999669 |

| Location | 8,421,632 – 8,421,742 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 96.44 |
| Mean single sequence MFE | -30.96 |
| Consensus MFE | -26.74 |
| Energy contribution | -27.18 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.949728 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
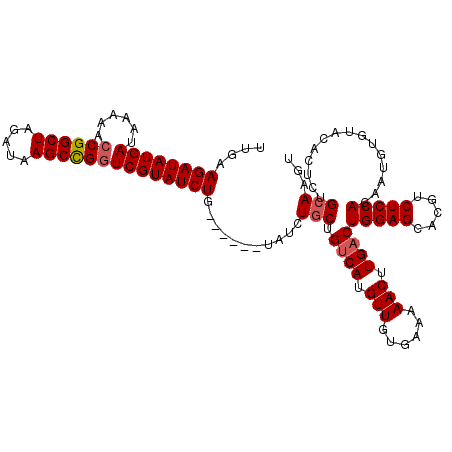
>2L_DroMel_CAF1 8421632 110 + 22407834 UUGAAGAUAUGAUAAAAACCGGCUAGAUAAGCCGGUCGUAUCUG------UAUCUGCUGUCAUGUUGUGAAAAACUUGACUGGAGCACGUCUCCACAAUGUGUACACUCUGCAAGU ....((((((((((...(((((((.....)))))))..)))).)------)))))((.((((.(((......))).))))(((((.....)))))...............)).... ( -32.70) >DroSec_CAF1 19802 110 + 1 UUGAAGAUAUGAUAAAAACCGGCUAGAUAAGCCGGUCGUAUCUG------UAUCUACUGUCAUGUUGUGAAAAACUUGACUGGAGCACGUCUCCACAAUGUGUACACUCUGCAAGU ....((((((((((...(((((((.....)))))))..)))).)------)))))...((((.(((......))).))))(((((.....))))).....((((.....))))... ( -31.70) >DroSim_CAF1 19620 116 + 1 UUGAAGAUAUGAUAAAAACCGGCUAGAUAAGCCGUUCGUAUCUGUAUCUGUAUCUGCUGUCAUGUUGUGAAAAACUUGACUGGAGCACGUCUCCACAAUGUGUACACUCUGCAAGU ....((((((((((..(((.((((.....)))))))..)))).)))))).....(((.((((.(((......))).))))(((((.....)))))...............)))... ( -28.80) >DroEre_CAF1 19956 110 + 1 UUGAAGAUAUGAUAAAAACCGGCUAGAUAAGCCGGUCGUAUCUG------UAUCUGCUGCCAUGUUGUGAAAAACUUGACUGGAGCACGUCUCCACAAUGUGUACACUCUGCAAGU ....((((((((((...(((((((.....)))))))..)))).)------)))))(((((...((.(((....(((((..(((((.....)))))))).)).)))))...)).))) ( -31.10) >DroYak_CAF1 20005 110 + 1 UUGAAGAUAUGAUAAAAACCGGCUAGAUAAGCUGGUCGUAUCUG------UAUCUGCUGUCAUGUUGUGAAAAACUUGACUGGAGCACGUCUCCACAAUGUGUACACUCUGCAAGU ....((((((((((...(((((((.....)))))))..)))).)------)))))((.((((.(((......))).))))(((((.....)))))...............)).... ( -30.50) >consensus UUGAAGAUAUGAUAAAAACCGGCUAGAUAAGCCGGUCGUAUCUG______UAUCUGCUGUCAUGUUGUGAAAAACUUGACUGGAGCACGUCUCCACAAUGUGUACACUCUGCAAGU ....((((((((......((((((.....))))))))))))))...........(((.((((.(((......))).))))(((((.....)))))...............)))... (-26.74 = -27.18 + 0.44)

| Location | 8,421,632 – 8,421,742 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 96.44 |
| Mean single sequence MFE | -31.14 |
| Consensus MFE | -29.16 |
| Energy contribution | -29.24 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.94 |
| SVM decision value | 3.86 |
| SVM RNA-class probability | 0.999669 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8421632 110 - 22407834 ACUUGCAGAGUGUACACAUUGUGGAGACGUGCUCCAGUCAAGUUUUUCACAACAUGACAGCAGAUA------CAGAUACGACCGGCUUAUCUAGCCGGUUUUUAUCAUAUCUUCAA ...(((..((((....)))).(((((.....)))))((((.(((......))).)))).)))((((------..((((.((((((((.....))))))))..)))).))))..... ( -32.90) >DroSec_CAF1 19802 110 - 1 ACUUGCAGAGUGUACACAUUGUGGAGACGUGCUCCAGUCAAGUUUUUCACAACAUGACAGUAGAUA------CAGAUACGACCGGCUUAUCUAGCCGGUUUUUAUCAUAUCUUCAA ...((.(((((((..((....(((((.....)))))((((.(((......))).)))).))..)))------).((((.((((((((.....))))))))..))))...))).)). ( -33.90) >DroSim_CAF1 19620 116 - 1 ACUUGCAGAGUGUACACAUUGUGGAGACGUGCUCCAGUCAAGUUUUUCACAACAUGACAGCAGAUACAGAUACAGAUACGAACGGCUUAUCUAGCCGGUUUUUAUCAUAUCUUCAA ..((((..((((....)))).(((((.....)))))((((.(((......))).)))).))))....(((((..((((.((((((((.....)))).)))).)))).))))).... ( -29.80) >DroEre_CAF1 19956 110 - 1 ACUUGCAGAGUGUACACAUUGUGGAGACGUGCUCCAGUCAAGUUUUUCACAACAUGGCAGCAGAUA------CAGAUACGACCGGCUUAUCUAGCCGGUUUUUAUCAUAUCUUCAA ...(((..((((....)))).(((((.....)))))((((.(((......))).)))).)))((((------..((((.((((((((.....))))))))..)))).))))..... ( -32.30) >DroYak_CAF1 20005 110 - 1 ACUUGCAGAGUGUACACAUUGUGGAGACGUGCUCCAGUCAAGUUUUUCACAACAUGACAGCAGAUA------CAGAUACGACCAGCUUAUCUAGCCGGUUUUUAUCAUAUCUUCAA ...(((..((((....)))).(((((.....)))))((((.(((......))).)))).)))((((------..((((.((((.(((.....))).))))..)))).))))..... ( -26.80) >consensus ACUUGCAGAGUGUACACAUUGUGGAGACGUGCUCCAGUCAAGUUUUUCACAACAUGACAGCAGAUA______CAGAUACGACCGGCUUAUCUAGCCGGUUUUUAUCAUAUCUUCAA ..((((..((((....)))).(((((.....)))))((((.(((......))).)))).))))...........((((.((((((((.....))))))))..)))).......... (-29.16 = -29.24 + 0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:39:45 2006