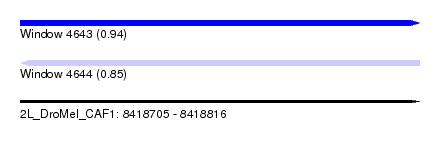
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,418,705 – 8,418,816 |
| Length | 111 |
| Max. P | 0.935505 |

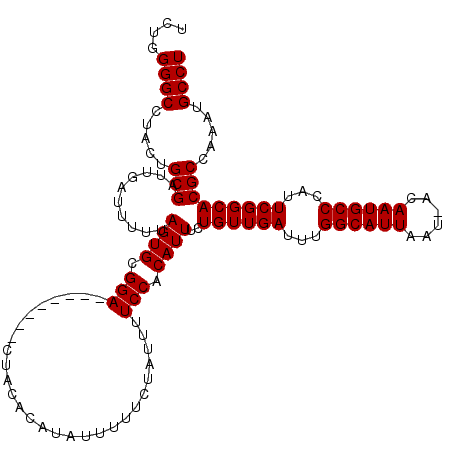
| Location | 8,418,705 – 8,418,816 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.60 |
| Mean single sequence MFE | -27.60 |
| Consensus MFE | -23.59 |
| Energy contribution | -23.99 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.935505 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8418705 111 + 22407834 AGGCAUUUGGCGUGCCGAAUGGGCAUUGU-AUUAAUGCCAAAUCAACAGAAAUGUGGAAAAUAGAAAUAUAUGUGUAG--------UCCGCACUAAAAAUCAAUCGCAGUAGGCCCCAGA .((.....(((.((((((.(((((((((.-..))))))).............((((((..(((.(......).)))..--------))))))........)).)))..))).)))))... ( -27.10) >DroSec_CAF1 16734 120 + 1 AGGCAUUUGGCGUGCCGAAUGGGCAUUGUUAUUAAUGCCAAAUCAACAGAAAUGUGGAAAAUAGAAAAAUAUGUGUAGUCAGUCAGUCCGCACUAAAAAUCAAUCGCAGUAGGCCCCAGA .((.....(((.((((((.(((((((((....)))))))...((.(((....))).))..............(((..(.(.....).)..))).......)).)))..))).)))))... ( -27.90) >DroSim_CAF1 16572 112 + 1 AGGCAUUUGGCGUGCCGAAUGGGCAUUGUUAUUAAUGCCAAAUCAACAGAAAUGUGGAAAAUAGAAAAAUAUGUGUAG--------UCCGCACUAAAAAUCAAUCGCAGUAGGCCCCAGA .((.....(((.((((((.(((((((((....))))))).............((((((..(((.(......).)))..--------))))))........)).)))..))).)))))... ( -28.80) >DroEre_CAF1 17020 110 + 1 AGGCACUUGGCGUGCCGAAUAGGCAUUGU-AUAAAUGCCAAAUCAACAAAAAUGUGGAAAAUAGAAAAAUAUGUGUAG--------UCCGCACUAA-AAUCAAUCGCAGUAGGCCCCAGA .((.....(((.((((((...((((((..-...)))))).............((((((..(((.(......).)))..--------))))))....-......)))..))).)))))... ( -27.10) >DroYak_CAF1 16893 111 + 1 AGGCAUUUGGCGUGCCGAAUGGGCAUUGU-AUUAAUGCCAAAUCAACAGAAAUGUGGAAAAUAGAAAAAUAUGUGUAG--------UCCGCACUAAAAAUCAAUCGCAGUAGGCCCCAGA .((.....(((.((((((.(((((((((.-..))))))).............((((((..(((.(......).)))..--------))))))........)).)))..))).)))))... ( -27.10) >consensus AGGCAUUUGGCGUGCCGAAUGGGCAUUGU_AUUAAUGCCAAAUCAACAGAAAUGUGGAAAAUAGAAAAAUAUGUGUAG________UCCGCACUAAAAAUCAAUCGCAGUAGGCCCCAGA .((.....(((.((((((.(((((((((....))))))).............((((((..(((......)))..............))))))........)).)))..))).)))))... (-23.59 = -23.99 + 0.40)

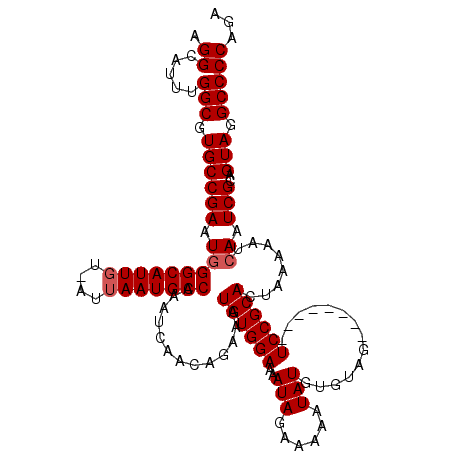
| Location | 8,418,705 – 8,418,816 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.60 |
| Mean single sequence MFE | -25.00 |
| Consensus MFE | -24.25 |
| Energy contribution | -24.25 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.847485 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8418705 111 - 22407834 UCUGGGGCCUACUGCGAUUGAUUUUUAGUGCGGA--------CUACACAUAUAUUUCUAUUUUCCACAUUUCUGUUGAUUUGGCAUUAAU-ACAAUGCCCAUUCGGCACGCCAAAUGCCU ....((((.....(((..........((((.(((--------....................))).))))..((((((...((((((...-..))))))...))))))))).....)))) ( -24.45) >DroSec_CAF1 16734 120 - 1 UCUGGGGCCUACUGCGAUUGAUUUUUAGUGCGGACUGACUGACUACACAUAUUUUUCUAUUUUCCACAUUUCUGUUGAUUUGGCAUUAAUAACAAUGCCCAUUCGGCACGCCAAAUGCCU ....((((.....(((((((.((.((((((((((..((..((.............))..)).))).((..((....))..))))))))).))))))(((.....))).))).....)))) ( -25.22) >DroSim_CAF1 16572 112 - 1 UCUGGGGCCUACUGCGAUUGAUUUUUAGUGCGGA--------CUACACAUAUUUUUCUAUUUUCCACAUUUCUGUUGAUUUGGCAUUAAUAACAAUGCCCAUUCGGCACGCCAAAUGCCU ....((((.....(((((((.((.((((((((((--------....................))).((..((....))..))))))))).))))))(((.....))).))).....)))) ( -24.75) >DroEre_CAF1 17020 110 - 1 UCUGGGGCCUACUGCGAUUGAUU-UUAGUGCGGA--------CUACACAUAUUUUUCUAUUUUCCACAUUUUUGUUGAUUUGGCAUUUAU-ACAAUGCCUAUUCGGCACGCCAAGUGCCU ....((((((...(((.......-..((((.(((--------....................))).))))..((((((...((((((...-..))))))...)))))))))..)).)))) ( -26.15) >DroYak_CAF1 16893 111 - 1 UCUGGGGCCUACUGCGAUUGAUUUUUAGUGCGGA--------CUACACAUAUUUUUCUAUUUUCCACAUUUCUGUUGAUUUGGCAUUAAU-ACAAUGCCCAUUCGGCACGCCAAAUGCCU ....((((.....(((..........((((.(((--------....................))).))))..((((((...((((((...-..))))))...))))))))).....)))) ( -24.45) >consensus UCUGGGGCCUACUGCGAUUGAUUUUUAGUGCGGA________CUACACAUAUUUUUCUAUUUUCCACAUUUCUGUUGAUUUGGCAUUAAU_ACAAUGCCCAUUCGGCACGCCAAAUGCCU ....((((.....(((..........((((.(((............................))).))))..((((((...((((((......))))))...))))))))).....)))) (-24.25 = -24.25 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:39:42 2006