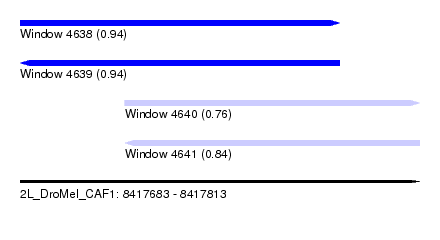
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,417,683 – 8,417,813 |
| Length | 130 |
| Max. P | 0.939909 |

| Location | 8,417,683 – 8,417,787 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 79.65 |
| Mean single sequence MFE | -27.95 |
| Consensus MFE | -18.50 |
| Energy contribution | -18.70 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939372 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
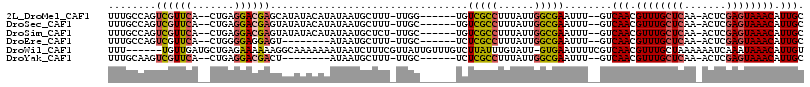
>2L_DroMel_CAF1 8417683 104 + 22407834 UUUGCCAGUCGUUCA--CUGAGGACGAGCAUAUACAUAUAAUGCUUU-UUGG------UGUCGCCUUUAUUGGCGAAUUU--GUCAACGUUUGCUCAA-ACUCGAGUAAACAUUGC ...(((((((((((.--....))))))((((.........))))...-))))------).(((((......)))))....--..(((.((((((((..-....)))))))).))). ( -32.00) >DroSec_CAF1 15722 104 + 1 UUUGCCAGUCGUUCA--CUGAGGACGAGUAUAUACAUAUAAUGCUUU-UUGC------UGUCGCCUUUAUUGGCGAAUUU--GUCAACGUUUGCUCAA-ACUCGAGUAAACAUUGC .....((((.....)--)))..(((((((.............((...-..))------..(((((......)))))))))--)))...((((((((..-....))))))))..... ( -28.90) >DroSim_CAF1 15568 104 + 1 UUUGCCAGUCGUUCA--CUGAGGACGAGUAUAUACAUAUAAUGCUCU-UUGC------UGUCGCCUUUAUUGGCGAAUUU--GUCAACGUUUGCUCAA-ACUCGAGUAAACAUUGC .....((((.((((.--....))))((((((.........)))))).-..))------))(((((......)))))....--..(((.((((((((..-....)))))))).))). ( -29.50) >DroEre_CAF1 16052 96 + 1 UUUGCCAGUCGUUCA--CUGGGGAGGAGU--------AUAAUGCUUU-UUGC------UCUCGCCUUUAUUGGCGAAUUU--GUCAACGUUUGCUCAA-ACUCGAGUAAACAUUGC (((.(((((.....)--)))).)))((((--------(.((....))-.)))------))(((((......)))))....--..(((.((((((((..-....)))))))).))). ( -30.50) >DroWil_CAF1 35940 109 + 1 UUU------UGUUGAUGCUGAGAAAAAAGGCAAAAAAAUAAUCUUUCGUUAUUGUUUGUCUUAUUUGUAUU-GUGAAUUUUCGUCAACGUUUGCUAAAAAAUCAAAUAAACAUUGU ...------.((((((((..(.(((.((((((((..((((((.....)))))).)))))))).))).)...-))((....))))))))(((((.............)))))..... ( -15.72) >DroYak_CAF1 15872 96 + 1 UUUGCAAGUCGUUCA--CUGAGGACGACU--------AUAAUGCUUU-UUGC------UCUCGCCUUUAUUGGCGAAUUU--GUCAACGUUUGCUCAA-ACUCGAGUAAACAUUGC ...(((((((((((.--....))))))))--------....)))...-....------..(((((......)))))....--..(((.((((((((..-....)))))))).))). ( -31.10) >consensus UUUGCCAGUCGUUCA__CUGAGGACGAGUAUAUACAUAUAAUGCUUU_UUGC______UCUCGCCUUUAUUGGCGAAUUU__GUCAACGUUUGCUCAA_ACUCGAGUAAACAUUGC ........((((((.......)))))).................................(((((......)))))........(((.((((((((.......)))))))).))). (-18.50 = -18.70 + 0.20)
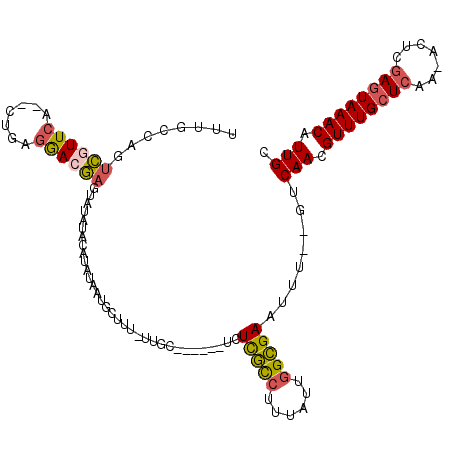

| Location | 8,417,683 – 8,417,787 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 79.65 |
| Mean single sequence MFE | -26.04 |
| Consensus MFE | -15.60 |
| Energy contribution | -15.85 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939909 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8417683 104 - 22407834 GCAAUGUUUACUCGAGU-UUGAGCAAACGUUGAC--AAAUUCGCCAAUAAAGGCGACA------CCAA-AAAGCAUUAUAUGUAUAUGCUCGUCCUCAG--UGAACGACUGGCAAA ((..((((((((.(((.-.((((((.........--....(((((......)))))..------....-...((((...))))...))))))..)))))--))))))....))... ( -31.80) >DroSec_CAF1 15722 104 - 1 GCAAUGUUUACUCGAGU-UUGAGCAAACGUUGAC--AAAUUCGCCAAUAAAGGCGACA------GCAA-AAAGCAUUAUAUGUAUAUACUCGUCCUCAG--UGAACGACUGGCAAA ((..((((((((.(((.-.((((.....((((..--....(((((......)))))))------))..-...((((...)))).....))))..)))))--))))))....))... ( -28.70) >DroSim_CAF1 15568 104 - 1 GCAAUGUUUACUCGAGU-UUGAGCAAACGUUGAC--AAAUUCGCCAAUAAAGGCGACA------GCAA-AGAGCAUUAUAUGUAUAUACUCGUCCUCAG--UGAACGACUGGCAAA ((..((((((((.(((.-.((((.....((((..--....(((((......)))))))------))..-...((((...)))).....))))..)))))--))))))....))... ( -28.70) >DroEre_CAF1 16052 96 - 1 GCAAUGUUUACUCGAGU-UUGAGCAAACGUUGAC--AAAUUCGCCAAUAAAGGCGAGA------GCAA-AAAGCAUUAU--------ACUCCUCCCCAG--UGAACGACUGGCAAA .((((((((.((((...-.)))).))))))))..--...((((((......)))))).------((..-...)).....--------........((((--(.....))))).... ( -26.10) >DroWil_CAF1 35940 109 - 1 ACAAUGUUUAUUUGAUUUUUUAGCAAACGUUGACGAAAAUUCAC-AAUACAAAUAAGACAAACAAUAACGAAAGAUUAUUUUUUUGCCUUUUUUCUCAGCAUCAACA------AAA .....((((..((((....)))).))))(((((.(((((.....-.........(((.((((.((((((....).)))))..)))).))))))))))))).......------... ( -12.13) >DroYak_CAF1 15872 96 - 1 GCAAUGUUUACUCGAGU-UUGAGCAAACGUUGAC--AAAUUCGCCAAUAAAGGCGAGA------GCAA-AAAGCAUUAU--------AGUCGUCCUCAG--UGAACGACUUGCAAA .((((((((.((((...-.)))).))))))))..--...((((((......)))))).------....-...(((....--------((((((.(....--.).)))))))))... ( -28.80) >consensus GCAAUGUUUACUCGAGU_UUGAGCAAACGUUGAC__AAAUUCGCCAAUAAAGGCGACA______GCAA_AAAGCAUUAUAUGUAUAUACUCGUCCUCAG__UGAACGACUGGCAAA .((((((((.(((((...))))).))))))))........(((((......)))))............................................................ (-15.60 = -15.85 + 0.25)

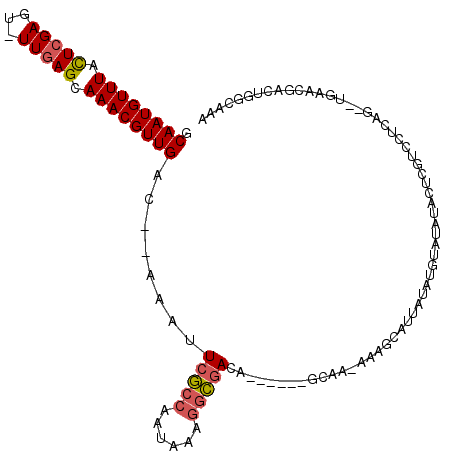
| Location | 8,417,717 – 8,417,813 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 79.72 |
| Mean single sequence MFE | -27.65 |
| Consensus MFE | -19.39 |
| Energy contribution | -18.95 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.764161 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8417717 96 + 22407834 UAUAAUGCUUU-UUGG------UGUCGCCUUUAUUGGCGAAUUU--GUCAACGUUUGCUCAA-ACUCGAGUAAACAUUGCAUUGCGAUACGGUGUGGAGGGACUGG--- ..((((((...-(((.------.((((((......)))))...)--..))).((((((((..-....))))))))...)))))).....((((........)))).--- ( -27.40) >DroSec_CAF1 15756 96 + 1 UAUAAUGCUUU-UUGC------UGUCGCCUUUAUUGGCGAAUUU--GUCAACGUUUGCUCAA-ACUCGAGUAAACAUUGCAUUGCGAUACGGAGUGGAGGGACUGG--- .......((((-(..(------(.(((((......)))))...(--(.(((.((((((((..-....)))))))).)))))...........))..))))).....--- ( -28.40) >DroSim_CAF1 15602 96 + 1 UAUAAUGCUCU-UUGC------UGUCGCCUUUAUUGGCGAAUUU--GUCAACGUUUGCUCAA-ACUCGAGUAAACAUUGCAUUGCGAUACGGAGUGGAGGGACUGG--- .......((((-(..(------(.(((((......)))))...(--(.(((.((((((((..-....)))))))).)))))...........))..))))).....--- ( -30.90) >DroEre_CAF1 16079 95 + 1 -AUAAUGCUUU-UUGC------UCUCGCCUUUAUUGGCGAAUUU--GUCAACGUUUGCUCAA-ACUCGAGUAAACAUUGCAUUGCGAUACGGCGGGGAGGGACCGG--- -....((((..-((((------..(((((......)))))...(--(.(((.((((((((..-....)))))))).)))))..))))...))))((......))..--- ( -29.70) >DroWil_CAF1 35970 108 + 1 AAUAAUCUUUCGUUAUUGUUUGUCUUAUUUGUAUU-GUGAAUUUUCGUCAACGUUUGCUAAAAAAUCAAAUAAACAUUGUGGGGCAGUCAAAUUCAGUCAGACUGAGAG ......((((((.....(((((.((.(((((.(((-((.....((((.(((.(((((.............))))).)))))))))))))))))..)).))))))))))) ( -16.72) >DroYak_CAF1 15899 95 + 1 -AUAAUGCUUU-UUGC------UCUCGCCUUUAUUGGCGAAUUU--GUCAACGUUUGCUCAA-ACUCGAGUAAACAUUGCAUUGUGAAUGCGGGCGGAGGGACUGG--- -......((((-((((------(((((((......)))))....--......((((((((..-....))))))))...((((.....)))))))))))))).....--- ( -32.80) >consensus UAUAAUGCUUU_UUGC______UCUCGCCUUUAUUGGCGAAUUU__GUCAACGUUUGCUCAA_ACUCGAGUAAACAUUGCAUUGCGAUACGGAGUGGAGGGACUGG___ ........................(((((......)))))......(((...((((((((.......))))))))(((((...)))))............)))...... (-19.39 = -18.95 + -0.44)



| Location | 8,417,717 – 8,417,813 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 79.72 |
| Mean single sequence MFE | -21.72 |
| Consensus MFE | -15.60 |
| Energy contribution | -15.85 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.844503 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8417717 96 - 22407834 ---CCAGUCCCUCCACACCGUAUCGCAAUGCAAUGUUUACUCGAGU-UUGAGCAAACGUUGAC--AAAUUCGCCAAUAAAGGCGACA------CCAA-AAAGCAUUAUA ---.....................((..((((((((((.((((...-.)))).)))))))).)--)...(((((......)))))..------....-...))...... ( -22.10) >DroSec_CAF1 15756 96 - 1 ---CCAGUCCCUCCACUCCGUAUCGCAAUGCAAUGUUUACUCGAGU-UUGAGCAAACGUUGAC--AAAUUCGCCAAUAAAGGCGACA------GCAA-AAAGCAUUAUA ---.....................((..((((((((((.((((...-.)))).)))))))).)--)...(((((......)))))..------))..-........... ( -22.10) >DroSim_CAF1 15602 96 - 1 ---CCAGUCCCUCCACUCCGUAUCGCAAUGCAAUGUUUACUCGAGU-UUGAGCAAACGUUGAC--AAAUUCGCCAAUAAAGGCGACA------GCAA-AGAGCAUUAUA ---.....................((..((((((((((.((((...-.)))).)))))))).)--)...(((((......)))))..------))..-........... ( -22.10) >DroEre_CAF1 16079 95 - 1 ---CCGGUCCCUCCCCGCCGUAUCGCAAUGCAAUGUUUACUCGAGU-UUGAGCAAACGUUGAC--AAAUUCGCCAAUAAAGGCGAGA------GCAA-AAAGCAUUAU- ---.((((........))))....((..((((((((((.((((...-.)))).)))))))).)--)..((((((......)))))).------))..-..........- ( -26.70) >DroWil_CAF1 35970 108 - 1 CUCUCAGUCUGACUGAAUUUGACUGCCCCACAAUGUUUAUUUGAUUUUUUAGCAAACGUUGACGAAAAUUCAC-AAUACAAAUAAGACAAACAAUAACGAAAGAUUAUU ....(((((...........)))))........((((((((((.....(((((....))))).((....))..-....))))).)))))...((((((....).))))) ( -13.60) >DroYak_CAF1 15899 95 - 1 ---CCAGUCCCUCCGCCCGCAUUCACAAUGCAAUGUUUACUCGAGU-UUGAGCAAACGUUGAC--AAAUUCGCCAAUAAAGGCGAGA------GCAA-AAAGCAUUAU- ---...(((.........((((.....))))(((((((.((((...-.)))).))))))))))--...((((((......)))))).------((..-...)).....- ( -23.70) >consensus ___CCAGUCCCUCCACACCGUAUCGCAAUGCAAUGUUUACUCGAGU_UUGAGCAAACGUUGAC__AAAUUCGCCAAUAAAGGCGACA______GCAA_AAAGCAUUAUA ..............................((((((((.(((((...))))).))))))))........(((((......)))))........................ (-15.60 = -15.85 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:39:39 2006