| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,403,720 – 8,403,814 |
| Length | 94 |
| Max. P | 0.988179 |

| Location | 8,403,720 – 8,403,814 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 76.09 |
| Mean single sequence MFE | -41.15 |
| Consensus MFE | -31.02 |
| Energy contribution | -32.50 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.46 |
| Structure conservation index | 0.75 |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.986992 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
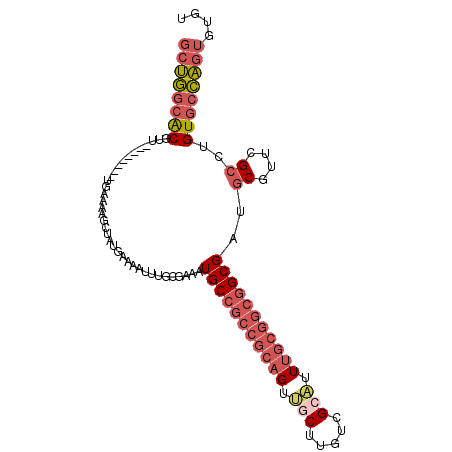
>2L_DroMel_CAF1 8403720 94 + 22407834 GCUGGCGCGUUU-------UGAAAAGCUAAGAAAAUUUGGGAAAUGCCGCCGCAGU--CUUGUCGCAUUUGCGGCGGCGAUGCGUUCGCCUGUGCCAGUGUGU ((((((((((((-------....))))...........(((((.(((((((((((.--..........))))))))))).....))).)).)))))))).... ( -38.00) >DroVir_CAF1 3916 89 + 1 ACAACAACCCGAAGCCGUGCGUACAAU--UU-AGAUUUGUGAAAUGCCGUCGCAGUCUCGAGAUGCGUU-------GCGCUGCUU-UGAUUGUAUUU---UGC ...((((..((((((.(((((.((.((--((-(((((.((((.......)))))))))..))))..)))-------)))).))))-)).))))....---... ( -23.60) >DroSec_CAF1 1839 95 + 1 GCUGGCACGUU--------UGAAAAGUUAUGAAAAUUUGCGAAAUGCCGCCGCAGUUGCUUGUCGCAUUUGCGGCGGCGAUGCGUUCGCCUGUGCCAGUGUGU ((((((((...--------....(((((.....)))))(((((.(((((((((((.(((.....))).))))))))))).....)))))..)))))))).... ( -45.10) >DroSim_CAF1 1816 95 + 1 GCUGGCACGUU--------UGAGAAGUUAUGAAAAUUUGCGAAAUGCCGCCGCAGUUGCUUGUCGCAUUUGCGGCGGCGAUGCGUUCGCCUGUGCCAGUGUGU ((((((((...--------....(((((.....)))))(((((.(((((((((((.(((.....))).))))))))))).....)))))..)))))))).... ( -45.20) >DroEre_CAF1 1890 94 + 1 GCUGGCGCGUG--------UGAAAAGCUCCGAAAAU-UGUGAAAUGCCGCCGCAGUUGCUCGUCGCAUUUGCGGCGGCGAUGCGUUCGCCUGUGCCAGUGUGU (((((((((.(--------((((..(((((((...)-)).))..(((((((((((.(((.....))).)))))))))))..)).))))).))))))))).... ( -47.20) >DroYak_CAF1 1887 94 + 1 GCUGGCGCGUU--------UGAAAAGCUUCGAAAAU-UGCGAAAUGCCGCCGCAGUUGCUCGUCGCAUUUGCGGCGGCGAUGCGUUCGCCUGUGCCAGUGUGU ((((((((.((--------((((....))))))...-.(((((.(((((((((((.(((.....))).))))))))))).....)))))..)))))))).... ( -47.80) >consensus GCUGGCACGUU________UGAAAAGCUAUGAAAAUUUGCGAAAUGCCGCCGCAGUUGCUUGUCGCAUUUGCGGCGGCGAUGCGUUCGCCUGUGCCAGUGUGU ((((((((....................................(((((((((((.(((.....))).)))))))))))..((....))..)))))))).... (-31.02 = -32.50 + 1.48)


| Location | 8,403,720 – 8,403,814 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 76.09 |
| Mean single sequence MFE | -34.27 |
| Consensus MFE | -25.66 |
| Energy contribution | -27.22 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.37 |
| Structure conservation index | 0.75 |
| SVM decision value | 2.11 |
| SVM RNA-class probability | 0.988179 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
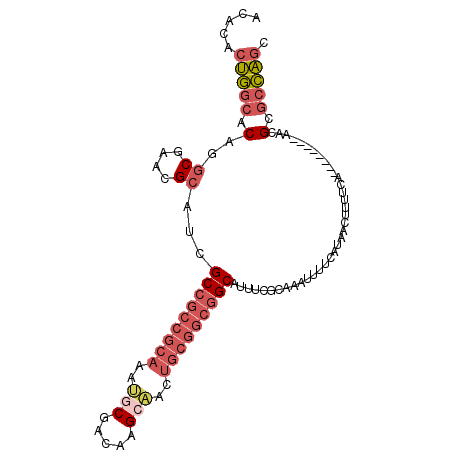
>2L_DroMel_CAF1 8403720 94 - 22407834 ACACACUGGCACAGGCGAACGCAUCGCCGCCGCAAAUGCGACAAG--ACUGCGGCGGCAUUUCCCAAAUUUUCUUAGCUUUUCA-------AAACGCGCCAGC .....(((((.(..((....))...(((((((((....(.....)--..)))))))))..........................-------....).))))). ( -30.80) >DroVir_CAF1 3916 89 - 1 GCA---AAAUACAAUCA-AAGCAGCGC-------AACGCAUCUCGAGACUGCGACGGCAUUUCACAAAUCU-AA--AUUGUACGCACGGCUUCGGGUUGUUGU ...---....((((((.-((((.((((-------..((((((....)).))))...)).....((((....-..--.))))..))...))))..))))))... ( -19.10) >DroSec_CAF1 1839 95 - 1 ACACACUGGCACAGGCGAACGCAUCGCCGCCGCAAAUGCGACAAGCAACUGCGGCGGCAUUUCGCAAAUUUUCAUAACUUUUCA--------AACGUGCCAGC .....(((((((..((....))...(((((((((..(((.....)))..)))))))))..........................--------...))))))). ( -40.20) >DroSim_CAF1 1816 95 - 1 ACACACUGGCACAGGCGAACGCAUCGCCGCCGCAAAUGCGACAAGCAACUGCGGCGGCAUUUCGCAAAUUUUCAUAACUUCUCA--------AACGUGCCAGC .....(((((((..((....))...(((((((((..(((.....)))..)))))))))..........................--------...))))))). ( -40.20) >DroEre_CAF1 1890 94 - 1 ACACACUGGCACAGGCGAACGCAUCGCCGCCGCAAAUGCGACGAGCAACUGCGGCGGCAUUUCACA-AUUUUCGGAGCUUUUCA--------CACGCGCCAGC .....(((((.(..(.(((.((...(((((((((..(((.....)))..)))))))))..(((...-......)))))..))).--------)..).))))). ( -36.60) >DroYak_CAF1 1887 94 - 1 ACACACUGGCACAGGCGAACGCAUCGCCGCCGCAAAUGCGACGAGCAACUGCGGCGGCAUUUCGCA-AUUUUCGAAGCUUUUCA--------AACGCGCCAGC .....(((((....(((((...((.(((((((((..(((.....)))..)))))))))))))))).-.........((......--------...))))))). ( -38.70) >consensus ACACACUGGCACAGGCGAACGCAUCGCCGCCGCAAAUGCGACAAGCAACUGCGGCGGCAUUUCGCAAAUUUUCAUAACUUUUCA________AACGCGCCAGC .....(((((.(..((....))...(((((((((..(((.....)))..))))))))).....................................).))))). (-25.66 = -27.22 + 1.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:39:33 2006