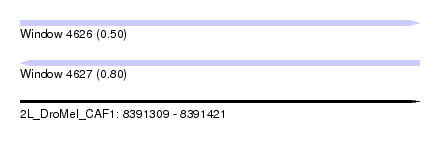
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,391,309 – 8,391,421 |
| Length | 112 |
| Max. P | 0.804998 |

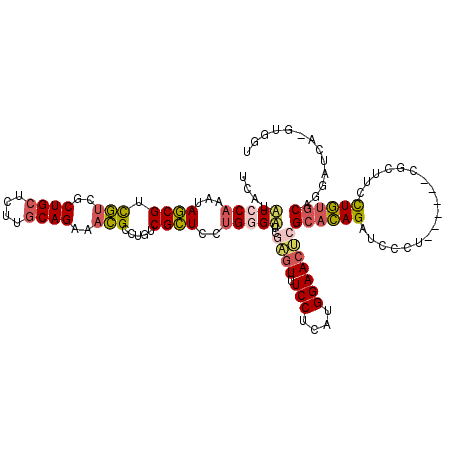
| Location | 8,391,309 – 8,391,421 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 81.69 |
| Mean single sequence MFE | -37.38 |
| Consensus MFE | -22.55 |
| Energy contribution | -22.89 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.60 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8391309 112 + 22407834 UCAUAUCCAAAUAGCGUCGUCGCUGCUCUUGCAGGAACGCCUGUCGCUCCUGGGUUCGAGUUUCCUCAUGGAACCCGCACAGAUCCCU------CGCUUUCUGUGCAGGAUCC-GUGGU ((..(((((...((((.(((..((((....))))..))).....))))..)))))..))......(((((((.((.(((((((.....------.....))))))).)).)))-)))). ( -40.70) >DroSec_CAF1 12883 112 + 1 CCAUAUCCAAAUAGCGUCGUCGCUGCUCUUGCAGGAACGCCUGUCGCUCCUGGGUUCGAGUUUCCUCAUGGAACUCGCACAGAUCCCU------CGCUUUCUGUGCAAGAUCG-GUGGU (((((((((...((((.(((..((((....))))..))).....))))..)))))(((((((.((....)))))))(((((((.....------.....)))))))..))...-)))). ( -38.50) >DroSim_CAF1 11965 112 + 1 CCAUAUCCAAAUAGCGUCGUCGCUGCUCUUGCAGGAACGCCUGUCGCUCCUGGGUUCGAGUUUCCUCAUGGAACUCGCACAGAUCCCU------CGCUUUCUGUGCAGGAUCG-GUGGU .............((((..((.((((....))))))))))(..(((.((((......(((((.((....)))))))(((((((.....------.....))))))))))).))-)..). ( -40.40) >DroEre_CAF1 12989 112 + 1 UCAUAUCCAAAUAGCGUCGUCGCUGCGCUUGCAGAAACGCCUGACGCUCCUGUGAUCUAGUUUCCUCAUGGAACUCGCACAGAUCCCU------CGCUUCCUGUGCAGGAUCA-GUGGC ............((((((((..((((....))))..))....))))))((..((((((((((.((....)))))).((((((......------......)))))).))))))-..)). ( -34.00) >DroYak_CAF1 13061 112 + 1 UCAUUUCCAAAUAGCGUCGUCGCUGCUCCUGCAGAAACGCCUUUCGCUCCUGGGAUCGAGUUUCCUCAUGGAACUCGCACAGAUCCCU------CACUUCCUGUGCAGGAUCA-GGAGC .............((((.....((((....))))..)))).....((((((((..(((((((.((....)))))))((((((......------......)))))).)).)))-))))) ( -39.80) >DroAna_CAF1 14393 119 + 1 UCAUUUCCAGGUAACGAUUCCGCUGAUCAACCAGAAAGGCUUGACGUUCCAGUGACCUGGUCUCCUCAUGGAAACUGCAAAGUUCCUUUGAGGUCGAUUCUUUGACCGAAUCCUGUGGU ......((((((........(((((.((((((.....)).)))).....)))))))))))...((.((.(((.....(((((...))))).((((((....))))))...))))).)). ( -30.90) >consensus UCAUAUCCAAAUAGCGUCGUCGCUGCUCUUGCAGAAACGCCUGUCGCUCCUGGGAUCGAGUUUCCUCAUGGAACUCGCACAGAUCCCU______CGCUUCCUGUGCAGGAUCA_GUGGU ....(((((...((((.(((..((((....))))..))).....))))..)))))..((((.(((....)))))))((((((..................))))))............. (-22.55 = -22.89 + 0.34)

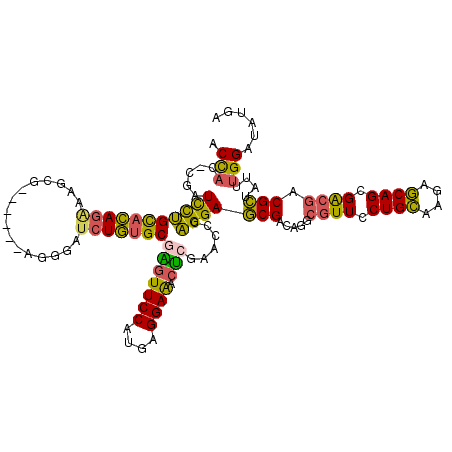
| Location | 8,391,309 – 8,391,421 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 81.69 |
| Mean single sequence MFE | -40.98 |
| Consensus MFE | -28.34 |
| Energy contribution | -27.37 |
| Covariance contribution | -0.96 |
| Combinations/Pair | 1.41 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.804998 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8391309 112 - 22407834 ACCAC-GGAUCCUGCACAGAAAGCG------AGGGAUCUGUGCGGGUUCCAUGAGGAAACUCGAACCCAGGAGCGACAGGCGUUCCUGCAAGAGCAGCGACGACGCUAUUUGGAUAUGA .....-(((.((((((((((.....------.....)))))))))).))).((((....))))...(((((((((.....((((.((((....)))).)))).)))).)))))...... ( -46.60) >DroSec_CAF1 12883 112 - 1 ACCAC-CGAUCUUGCACAGAAAGCG------AGGGAUCUGUGCGAGUUCCAUGAGGAAACUCGAACCCAGGAGCGACAGGCGUUCCUGCAAGAGCAGCGACGACGCUAUUUGGAUAUGG .(((.-.((.((((((((((.....------.....)))))))))).))..((((....))))...(((((((((.....((((.((((....)))).)))).)))).)))))...))) ( -41.60) >DroSim_CAF1 11965 112 - 1 ACCAC-CGAUCCUGCACAGAAAGCG------AGGGAUCUGUGCGAGUUCCAUGAGGAAACUCGAACCCAGGAGCGACAGGCGUUCCUGCAAGAGCAGCGACGACGCUAUUUGGAUAUGG .(((.-..((((((((((((.....------.....)))))))).((((..((((....))))....((((((((.....))))))))...))))((((....))))....)))).))) ( -41.60) >DroEre_CAF1 12989 112 - 1 GCCAC-UGAUCCUGCACAGGAAGCG------AGGGAUCUGUGCGAGUUCCAUGAGGAAACUAGAUCACAGGAGCGUCAGGCGUUUCUGCAAGCGCAGCGACGACGCUAUUUGGAUAUGA .(((.-...((((....))))((((------..(...((((((.((((((....)).))))......((((((((.....))))))))...))))))...)..))))...)))...... ( -35.10) >DroYak_CAF1 13061 112 - 1 GCUCC-UGAUCCUGCACAGGAAGUG------AGGGAUCUGUGCGAGUUCCAUGAGGAAACUCGAUCCCAGGAGCGAAAGGCGUUUCUGCAGGAGCAGCGACGACGCUAUUUGGAAAUGA (((((-((.((((....))))....------.((((((.(((.(....))))(((....)))))))))(((((((.....))))))).)))))))((((....))))............ ( -45.10) >DroAna_CAF1 14393 119 - 1 ACCACAGGAUUCGGUCAAAGAAUCGACCUCAAAGGAACUUUGCAGUUUCCAUGAGGAGACCAGGUCACUGGAACGUCAAGCCUUUCUGGUUGAUCAGCGGAAUCGUUACCUGGAAAUGA ....((((...(((((.....(((((((..(((((.((.((.((((..((.((.(....)))))..)))).)).))....)))))..))))))).....).))))...))))....... ( -35.90) >consensus ACCAC_CGAUCCUGCACAGAAAGCG______AGGGAUCUGUGCGAGUUCCAUGAGGAAACUCGAACCCAGGAGCGACAGGCGUUCCUGCAAGAGCAGCGACGACGCUAUUUGGAUAUGA .(((.....(((((((((((................)))))))(((((((....)))).)))......))))(((.....((((.((((....)))).)))).)))....)))...... (-28.34 = -27.37 + -0.96)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:39:26 2006