
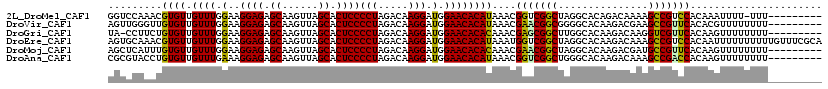
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,389,714 – 8,389,823 |
| Length | 109 |
| Max. P | 0.540572 |

| Location | 8,389,714 – 8,389,823 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 84.37 |
| Mean single sequence MFE | -32.33 |
| Consensus MFE | -27.10 |
| Energy contribution | -26.21 |
| Covariance contribution | -0.89 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.540572 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8389714 109 + 22407834 GGUCCAAACGUGUUGUUUGGAAGGAGAGCAAGUUAGCACUCCCCUAGACAAGGAUGGAACACAUAAACGGUCGGCUAGGCACAGACAAAAGCCGUCCACAAAUUUU-UUU--------- (.((((....(.((((((((..((((.((......)).)))).)))))))).).)))).)........((.(((((..(......)...))))).)).........-...--------- ( -31.00) >DroVir_CAF1 13377 110 + 1 AGUUGGGUUGUGUUGUUUGGAAGGAGAGCAAGUUAGCACUCCCCUAGACAAGGAUGGAACACAUAAACGAACGGCGGGGCACAAGACGAAGCCGUUCACACGUUUUUUUU--------- .(((....((((((((((((..((((.((......)).)))).)))))))........)))))..)))(((((((..(........)...))))))).............--------- ( -31.40) >DroGri_CAF1 11958 109 + 1 UA-CCUUCUGUGUUGUUUGGAAGGAGAGCAAGUUAGCACUCCCCUAGACAAGGAUGGAACACACAAACGAGCGGCUUGGCACAAGACAAGGUCGUUCACAAGUUUUUUUU--------- ..-((.(((...((((((((..((((.((......)).)))).))))))))))).))...........((((((((((........))).))))))).............--------- ( -30.90) >DroEre_CAF1 11319 119 + 1 AGUGCAAACGUGUUGUUUGGAAGGAGAGCAAGUUAGCACUCCCCUAGACAAGGAUGGAACACAUAAAUGGUCGGCUAGGCACAAGACAAAGCCGUCCACAAUUUUUUUUUUGUUUCGCA .(((.(((((((((((((((..((((.((......)).)))).)))))))........)))).....(((.(((((..((....).)..))))).))).............))))))). ( -34.30) >DroMoj_CAF1 12849 110 + 1 AGCUCAUUUGUGUUGUUUGGAAGGAGAGCAAGUUAGCACUCCCCUAGACAAGGAUGGAACACACAAACGAACGGCUAGGCACAAGACGAUGCCGUUCACAAGUUUUUUUU--------- ((((..(((((((.((((.(..((((.((......)).))))(((.....))).).))))))))))).(((((((..(........)...)))))))...))))......--------- ( -32.40) >DroAna_CAF1 11001 110 + 1 CGCGUACCUGUGUUGUUUGAAAGGAGAGCAAGUUAGCACUCCCCUAGACAAGGAUGGAACACAUAAACGGUCGGCUGGGCACAAGACAAAGCCGACCACAAGUUUUUUUU--------- .(.((.((..(.(((((((...((((.((......)).))))..))))))).)..)).)).)......((((((((..((....).)..)))))))).............--------- ( -34.00) >consensus AGUCCAAAUGUGUUGUUUGGAAGGAGAGCAAGUUAGCACUCCCCUAGACAAGGAUGGAACACAUAAACGAUCGGCUAGGCACAAGACAAAGCCGUCCACAAGUUUUUUUU_________ .........((((.((((.(..((((.((......)).))))(((.....))).).))))))))....(((((((...............)))))))...................... (-27.10 = -26.21 + -0.89)
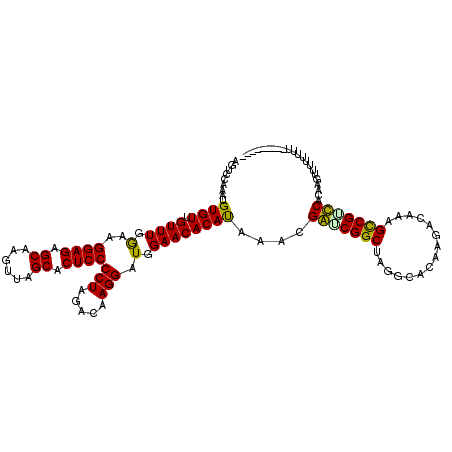
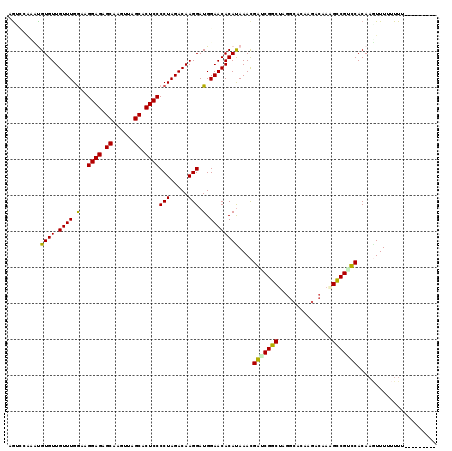
| Location | 8,389,714 – 8,389,823 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 84.37 |
| Mean single sequence MFE | -31.07 |
| Consensus MFE | -23.39 |
| Energy contribution | -22.69 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.75 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.528299 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8389714 109 - 22407834 ---------AAA-AAAAUUUGUGGACGGCUUUUGUCUGUGCCUAGCCGACCGUUUAUGUGUUCCAUCCUUGUCUAGGGGAGUGCUAACUUGCUCUCCUUCCAAACAACACGUUUGGACC ---------...-.........((.(((((...((....))..))))).))...............(((.....)))((((.((......)).)))).(((((((.....))))))).. ( -29.20) >DroVir_CAF1 13377 110 - 1 ---------AAAAAAAACGUGUGAACGGCUUCGUCUUGUGCCCCGCCGUUCGUUUAUGUGUUCCAUCCUUGUCUAGGGGAGUGCUAACUUGCUCUCCUUCCAAACAACACAACCCAACU ---------.........(((((((((((...((.....))...)))))))((((...........(((.....)))((((.((......)).))))....)))).))))......... ( -28.80) >DroGri_CAF1 11958 109 - 1 ---------AAAAAAAACUUGUGAACGACCUUGUCUUGUGCCAAGCCGCUCGUUUGUGUGUUCCAUCCUUGUCUAGGGGAGUGCUAACUUGCUCUCCUUCCAAACAACACAGAAGG-UA ---------.......(((((..(((((.((((.(....).))))....)))))..)(((((....(((.....)))((((.((......)).))))........)))))...)))-). ( -24.90) >DroEre_CAF1 11319 119 - 1 UGCGAAACAAAAAAAAAAUUGUGGACGGCUUUGUCUUGUGCCUAGCCGACCAUUUAUGUGUUCCAUCCUUGUCUAGGGGAGUGCUAACUUGCUCUCCUUCCAAACAACACGUUUGCACU (((.((((............((((.(((((..((.....))..))))).))))....(((((....(((.....)))((((.((......)).))))........)))))))))))).. ( -33.60) >DroMoj_CAF1 12849 110 - 1 ---------AAAAAAAACUUGUGAACGGCAUCGUCUUGUGCCUAGCCGUUCGUUUGUGUGUUCCAUCCUUGUCUAGGGGAGUGCUAACUUGCUCUCCUUCCAAACAACACAAAUGAGCU ---------.................(((((......))))).....(((((((((((((((....(((.....)))((((.((......)).)))).....))).)))))))))))). ( -34.50) >DroAna_CAF1 11001 110 - 1 ---------AAAAAAAACUUGUGGUCGGCUUUGUCUUGUGCCCAGCCGACCGUUUAUGUGUUCCAUCCUUGUCUAGGGGAGUGCUAACUUGCUCUCCUUUCAAACAACACAGGUACGCG ---------.............((((((((..((.....))..))))))))......((((.((....((((..(((((((.((......)).)))))))...))))....)).)))). ( -35.40) >consensus _________AAAAAAAACUUGUGAACGGCUUUGUCUUGUGCCUAGCCGACCGUUUAUGUGUUCCAUCCUUGUCUAGGGGAGUGCUAACUUGCUCUCCUUCCAAACAACACAAAUGAACU .....................(((.((((...............)))).)))....((((((....(((.....)))((((.((......)).))))........))))))........ (-23.39 = -22.69 + -0.69)

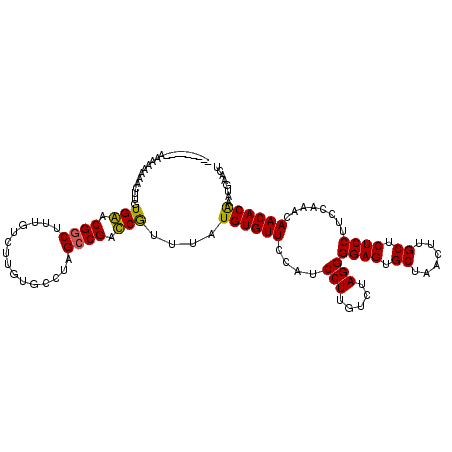
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:39:23 2006