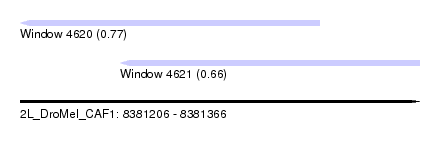
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,381,206 – 8,381,366 |
| Length | 160 |
| Max. P | 0.774882 |

| Location | 8,381,206 – 8,381,326 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.39 |
| Mean single sequence MFE | -46.22 |
| Consensus MFE | -32.92 |
| Energy contribution | -33.70 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.34 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.774882 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
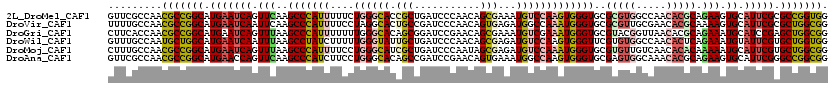
>2L_DroMel_CAF1 8381206 120 - 22407834 GUUCGCCAACGCCGGCAUGAAUCAGUUCAAGCCCAUUUUUCUGGGCACCGCUGAUCCCAACAGCGAAAUGUCCAAGUGGGUGCGCGUGGCCAACACGCAGAAGUGCAUUCGCGCCGGUGG .........(((((((.(((((((.(((..(((((((....((((((.(((((.......)))))...)))))))))))))..(((((.....))))).))).)).))))).))))))). ( -55.80) >DroVir_CAF1 2741 120 - 1 UUUUGCCAACGCCGGCAUGAAUCAAUUCAAGCCCAUUUUCCUAGGCACUGCCGAUCCCAACAGUGAGAUGGCCAAAUGGGUGCGCGUUGCGAACACGCAAAAGUGCAUUCGCGCUGGCGG .........(((((((.(((((........((((((((..(((..(((((..........)))))...)))..))))))))((((.(((((....)))))..))))))))).))))))). ( -46.70) >DroGri_CAF1 3346 120 - 1 CUUCACCAACGCCGGCAUGAAUCAGUUUAAGCCCAUUUUUUUGGGCACAGCGGAUCCGAACAGCGAAAUGUCGAAAUGGGUGCGUACGGUUAACACGCAGAAAUGCAUCCGAGCUGGCGG .........(((((((...((((.......(((((......)))))...(((.(((((.....(((....)))...))))).)))..)))).....(((....)))......))))))). ( -42.20) >DroWil_CAF1 2729 120 - 1 GUUUGCCAAUGCUGGCAUGAAUCAAUUUAAGCCUAUCUUUUUGGGUAUUGCUGAUCCCAACAGCGAGAUGUCCAAGUGGGUUCGUGUGGCCAACACUCAGAAAUGUAUUCGUGCUGGUGG ..........((..((((((((((.(((.(((((((....((((..(((((((.......))))..)))..))))))))))).((((.....))))...))).)).))))))))..)).. ( -37.30) >DroMoj_CAF1 2725 120 - 1 CUUUGCCAACGCCGGCAUGAAUCAGUUUAAGCCCAUUUUCCUGGGCAUCGCUGAUCCCAAUAGCGAGAUGUCCAAAUGGGUGCGUGUUGUCAACACACAAAAAUGCAUUCGUGCUGGCGG .........(((((((((((((((.(((..(((((((....((((((((((((.......))))))..)))))))))))))(.((((.....)))).).))).)).))))))))))))). ( -49.10) >DroAna_CAF1 2723 120 - 1 GUUCGCCAACGCCGGCAUGAACCAGUUCAAGCCCAUCUUCCUGGGCACAGCCGAUCCGAACAGUGAAAUGGCCAAGUGGGUGCGAGUGGCAAACACGCAGAAGUGCAUUCGGGCCGGCGG ...((((.....((((.((((....)))).(((((......)))))...))))................((((..(((..(((.....)))..)))(((....))).....)))))))). ( -46.20) >consensus GUUCGCCAACGCCGGCAUGAAUCAGUUCAAGCCCAUUUUCCUGGGCACAGCUGAUCCCAACAGCGAAAUGUCCAAAUGGGUGCGUGUGGCCAACACGCAGAAAUGCAUUCGCGCUGGCGG .........(((((((.(((((((.(((..(((((((....((((((.(((...........)))...)))))))))))))..(((((.....))))).))).)).))))).))))))). (-32.92 = -33.70 + 0.78)
| Location | 8,381,246 – 8,381,366 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.83 |
| Mean single sequence MFE | -38.48 |
| Consensus MFE | -27.25 |
| Energy contribution | -26.81 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.663198 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8381246 120 - 22407834 ACUCGUCCAGCACGAUUCCAUUGGACGAUCCCACGCUGUUGUUCGCCAACGCCGGCAUGAAUCAGUUCAAGCCCAUUUUUCUGGGCACCGCUGAUCCCAACAGCGAAAUGUCCAAGUGGG ..((((((((..........)))))))).((((((((((((...(((......)))....((((((....(((((......)))))...))))))..)))))))((....))...))))) ( -45.30) >DroVir_CAF1 2781 120 - 1 ACUCCUCCAGCACGAUACCGUUGGAUGAUCCCACCCUGCUUUUUGCCAACGCCGGCAUGAAUCAAUUCAAGCCCAUUUUCCUAGGCACUGCCGAUCCCAACAGUGAGAUGGCCAAAUGGG ..((.((((((........)))))).))........((.(((.((((......)))).))).)).......(((((((..(((..(((((..........)))))...)))..))))))) ( -30.40) >DroGri_CAF1 3386 120 - 1 ACUCCUCCAGCACGAUUCCGCUGGAUGAUCCAACGCUACUCUUCACCAACGCCGGCAUGAAUCAGUUUAAGCCCAUUUUUUUGGGCACAGCGGAUCCGAACAGCGAAAUGUCGAAAUGGG ......(((...((((..(((((...(((((...((((((.((((((......))..))))..)))....(((((......)))))..))))))))....)))))....))))...))). ( -36.10) >DroWil_CAF1 2769 120 - 1 AUUCGUCGAGUACAAUACCAUUAGAUGAUCCCACUCUAUUGUUUGCCAAUGCUGGCAUGAAUCAAUUUAAGCCUAUCUUUUUGGGUAUUGCUGAUCCCAACAGCGAGAUGUCCAAGUGGG ..(((((.(((........))).))))).((((((........(((((....)))))...(((.......(((((......))))).((((((.......))))))))).....)))))) ( -30.50) >DroMoj_CAF1 2765 120 - 1 ACUCGUCCAGCACCAUUCCGUUGGACGAUCCCACUCUACUCUUUGCCAACGCCGGCAUGAAUCAGUUUAAGCCCAUUUUCCUGGGCAUCGCUGAUCCCAAUAGCGAGAUGUCCAAAUGGG ..(((((((((........))))))))).((((......((..((((......)))).))(((.......(((((......))))).((((((.......))))))))).......)))) ( -40.20) >DroAna_CAF1 2763 120 - 1 ACUCGUCCAGCACGAUUCCCCUGGACGAUCCCACUUUGCUGUUCGCCAACGCCGGCAUGAACCAGUUCAAGCCCAUCUUCCUGGGCACAGCCGAUCCGAACAGUGAAAUGGCCAAGUGGG ..((((((((..........)))))))).(((((((.(((((((((...((.((((.((((....)))).(((((......)))))...))))...))....))).)))))).))))))) ( -48.40) >consensus ACUCGUCCAGCACGAUUCCGUUGGACGAUCCCACUCUACUGUUCGCCAACGCCGGCAUGAAUCAGUUCAAGCCCAUUUUCCUGGGCACAGCUGAUCCCAACAGCGAAAUGUCCAAAUGGG ..(((((((((........))))))))).((((........(((((......((((.((((....)))).(((((......)))))...)))).........))))).........)))) (-27.25 = -26.81 + -0.44)

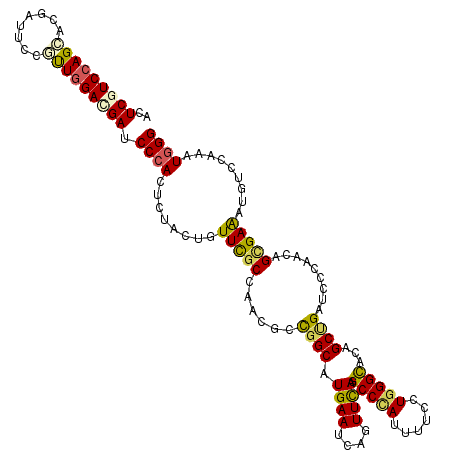
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:39:20 2006