
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,374,354 – 8,374,449 |
| Length | 95 |
| Max. P | 0.984644 |

| Location | 8,374,354 – 8,374,449 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 78.03 |
| Mean single sequence MFE | -27.88 |
| Consensus MFE | -17.86 |
| Energy contribution | -19.37 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.984644 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
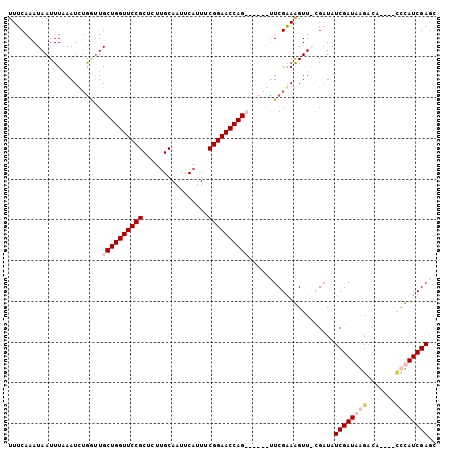
>2L_DroMel_CAF1 8374354 95 + 22407834 UUUCAAAUAAUUUAAAUCUGGUUGCUGGUUCCGCUCUUGCAAUUCAUUCCGGAACCAG------UUCGAAAGUUGCGAUAUCGAUAACACA----GCUAUCGACC .......................((((((((((....((.....))...)))))))))------)((((.(((((.(.((....)).).))----))).)))).. ( -23.60) >DroSim_CAF1 17528 95 + 1 UUUCAAAUAAUUUAAAUUGACUUGCUGGUUCCGCUUUUGCAAUUCAUUUCGGAACCAG------UUCGAAAGUUGCGUUAUCGAUAAGACA----GCUAUCGAGA .......................((((((((((....((.....))...)))))))))------)((((.(((((..(((....)))..))----))).)))).. ( -24.30) >DroEre_CAF1 17237 102 + 1 UUUCAAAUAAUUUAAAUCUGA-UGCUGGUUCCGCUCUUGCA-UUCUGUGCGGAACCAGUCGCAGCUCGGAGGUU-CGAUAUCGAUAGGACAUUCACUCAUCGAGC ..................((.-.(((((((((((.(.....-....).)))))))))))..))((((((((..(-((....)))...(.....).)))..))))) ( -30.80) >DroYak_CAF1 17143 104 + 1 UUUCAAAUAAUUUUAAUCUGGUUGCUGGUUCCGCAUUUGCAGUUCAUUUCGGAACCAGUAGCAGUUCGGAGGUU-UGAUAUCGAUAGGUCACUCACUCAUCGAGU ..((((((...((..(.(((.((((((((((((.(..((.....))..)))))))))))))))).)..)).)))-)))..(((((..((.....))..))))).. ( -32.80) >consensus UUUCAAAUAAUUUAAAUCUGGUUGCUGGUUCCGCUCUUGCAAUUCAUUUCGGAACCAG______UUCGAAAGUU_CGAUAUCGAUAAGACA____CCCAUCGAGC .......................((((((((((................)))))))))).....................((((((((.......)))))))).. (-17.86 = -19.37 + 1.50)


| Location | 8,374,354 – 8,374,449 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 78.03 |
| Mean single sequence MFE | -25.96 |
| Consensus MFE | -19.72 |
| Energy contribution | -19.84 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968791 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
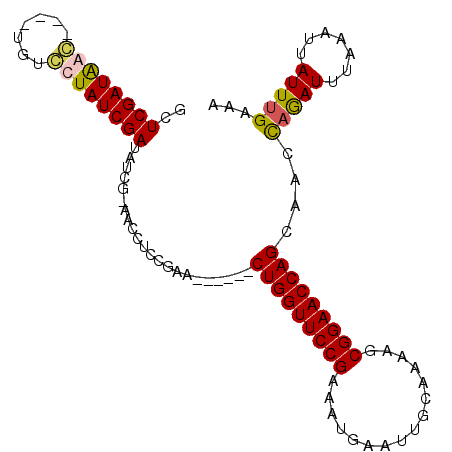
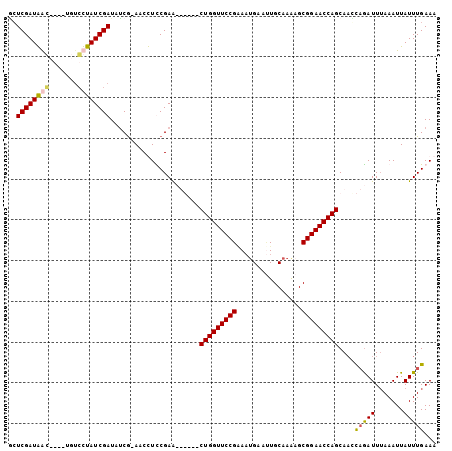
>2L_DroMel_CAF1 8374354 95 - 22407834 GGUCGAUAGC----UGUGUUAUCGAUAUCGCAACUUUCGAA------CUGGUUCCGGAAUGAAUUGCAAGAGCGGAACCAGCAACCAGAUUUAAAUUAUUUGAAA .(((((((((----...)))))))))...............------(((((((((...((.....))....)))))))))....(((((.......)))))... ( -24.40) >DroSim_CAF1 17528 95 - 1 UCUCGAUAGC----UGUCUUAUCGAUAACGCAACUUUCGAA------CUGGUUCCGAAAUGAAUUGCAAAAGCGGAACCAGCAAGUCAAUUUAAAUUAUUUGAAA ..((((.((.----(((.(((....))).))).)).)))).------(((((((((...((.....))....)))))))))(((((.(((....))))))))... ( -22.50) >DroEre_CAF1 17237 102 - 1 GCUCGAUGAGUGAAUGUCCUAUCGAUAUCG-AACCUCCGAGCUGCGACUGGUUCCGCACAGAA-UGCAAGAGCGGAACCAGCA-UCAGAUUUAAAUUAUUUGAAA (((((..(.((..(((((.....)))))..-.)))..)))))...(.((((((((((......-.......))))))))))).-((((((.......)))))).. ( -32.92) >DroYak_CAF1 17143 104 - 1 ACUCGAUGAGUGAGUGACCUAUCGAUAUCA-AACCUCCGAACUGCUACUGGUUCCGAAAUGAACUGCAAAUGCGGAACCAGCAACCAGAUUAAAAUUAUUUGAAA ..((((((.((.....)).))))))..(((-((......(((((...(((((((((...((.....))....)))))))))....))).)).......))))).. ( -24.02) >consensus GCUCGAUAAC____UGUCCUAUCGAUAUCG_AACCUCCGAA______CUGGUUCCGAAAUGAAUUGCAAAAGCGGAACCAGCAACCAGAUUUAAAUUAUUUGAAA ..((((((((.......))))))))......................(((((((((................)))))))))....(((((.......)))))... (-19.72 = -19.84 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:39:17 2006