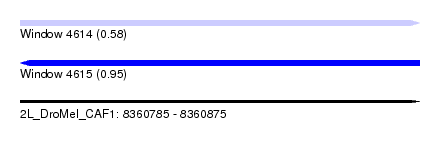
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,360,785 – 8,360,875 |
| Length | 90 |
| Max. P | 0.948847 |

| Location | 8,360,785 – 8,360,875 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
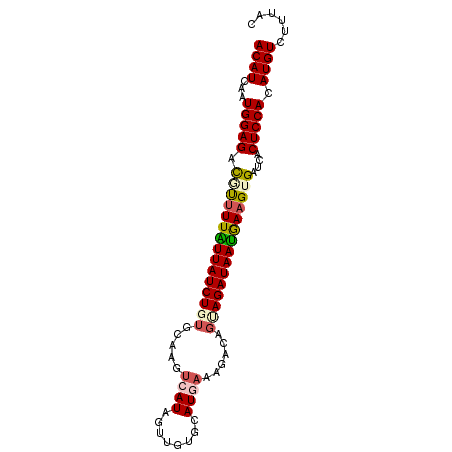
| Reading direction | forward |
| Mean pairwise identity | 88.22 |
| Mean single sequence MFE | -19.20 |
| Consensus MFE | -13.80 |
| Energy contribution | -13.60 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.580897 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8360785 90 + 22407834 GUAAAGACAUGUGGAGUGAUCACUUCAUUAUCUACUGUUUUUCAUGCACAACUAUGACUUGCACAGAUAAUAAAAUGUCUCCAUUGAUGU ....((((((..(((((....)))))(((((((..(((...(((((......)))))...))).)))))))...)))))).......... ( -19.10) >DroSec_CAF1 3443 90 + 1 GUAAAGACAUGUGGAGUGAUCACUUCAUUAUCUACUGUCUUUCAUGCACAAUUAUGACUUGCAAAGAUAAUAAAACGUCUCCAUUGAUGU (.(((((((.(((((((((.....))))..))))))))))))).((((...........))))...........(((((......))))) ( -20.10) >DroSim_CAF1 3461 90 + 1 GUGAAGACAUGUGGAGUGAUCACUUCAUUAUCUACUGUCUUUCAUGCACAACUAUGACUUGCAAAGAUAAUAAAACGUCUCCAUUGAUGU (((((((((.(((((((((.....))))..))))))))).)))))(((...........)))............(((((......))))) ( -20.20) >DroEre_CAF1 3426 87 + 1 GUA--GACAUUUGGAGUGAUCACUUCAUUAUCUGCUGUCUU-UAUACACAACUAUGACUUGCACAGAUAACAAAGAAUCUCCAAUGAUGU ...--.((((((((((......(((..(((((((.(((..(-((((......)))))...))))))))))..)))...)))))..))))) ( -19.10) >DroYak_CAF1 3478 90 + 1 GUAAAGACAUUUGGAGUGAUCACUUUUUUAUCUAAUGUCGUUGAUACACCACUAUGACUUGCACAGAUAAAAAUAAAUCUCCAUUGAUGU ......((((((((((........(((((((((...(((((((......))..)))))......))))))))).....)))))..))))) ( -17.52) >consensus GUAAAGACAUGUGGAGUGAUCACUUCAUUAUCUACUGUCUUUCAUGCACAACUAUGACUUGCACAGAUAAUAAAAAGUCUCCAUUGAUGU ......((((((((((.((.....))(((((((..(((...(((((......)))))...))).))))))).......))))))..)))) (-13.80 = -13.60 + -0.20)

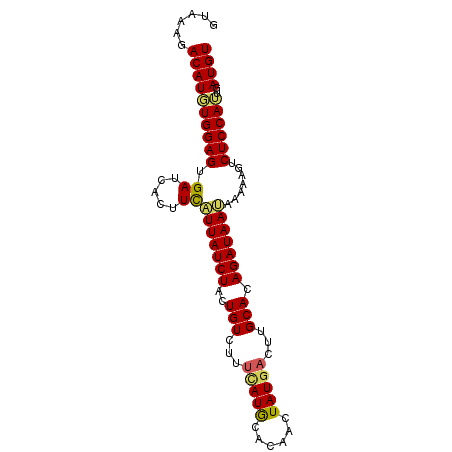
| Location | 8,360,785 – 8,360,875 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 88.22 |
| Mean single sequence MFE | -21.27 |
| Consensus MFE | -16.72 |
| Energy contribution | -17.36 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.948847 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8360785 90 - 22407834 ACAUCAAUGGAGACAUUUUAUUAUCUGUGCAAGUCAUAGUUGUGCAUGAAAAACAGUAGAUAAUGAAGUGAUCACUCCACAUGUCUUUAC ((((...(((((.((((((((((((((((....((((.(.....)))))....)).))))))))))))))....))))).))))...... ( -23.00) >DroSec_CAF1 3443 90 - 1 ACAUCAAUGGAGACGUUUUAUUAUCUUUGCAAGUCAUAAUUGUGCAUGAAAGACAGUAGAUAAUGAAGUGAUCACUCCACAUGUCUUUAC .......((((((((((((((((((((((....((((........))))....))).))))))))))(((.......)))))))))))). ( -20.90) >DroSim_CAF1 3461 90 - 1 ACAUCAAUGGAGACGUUUUAUUAUCUUUGCAAGUCAUAGUUGUGCAUGAAAGACAGUAGAUAAUGAAGUGAUCACUCCACAUGUCUUCAC .......((((((((((((((((((((((....((((.(.....)))))....))).))))))))))(((.......)))))))))))). ( -23.20) >DroEre_CAF1 3426 87 - 1 ACAUCAUUGGAGAUUCUUUGUUAUCUGUGCAAGUCAUAGUUGUGUAUA-AAGACAGCAGAUAAUGAAGUGAUCACUCCAAAUGUC--UAC ((((..((((((((((((..((((((((....(((.............-..))).))))))))..))).))))..))))))))).--... ( -23.26) >DroYak_CAF1 3478 90 - 1 ACAUCAAUGGAGAUUUAUUUUUAUCUGUGCAAGUCAUAGUGGUGUAUCAACGACAUUAGAUAAAAAAGUGAUCACUCCAAAUGUCUUUAC ((((...(((((..((((((((..(((((.....)))))((((((.......)))))).....))))))))...))))).))))...... ( -16.00) >consensus ACAUCAAUGGAGACGUUUUAUUAUCUGUGCAAGUCAUAGUUGUGCAUGAAAGACAGUAGAUAAUGAAGUGAUCACUCCACAUGUCUUUAC ((((...(((((.(((((((((((((((.....((((........))))......)))))))))))))))....))))).))))...... (-16.72 = -17.36 + 0.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:39:14 2006