| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,329,924 – 8,330,024 |
| Length | 100 |
| Max. P | 0.816947 |

| Location | 8,329,924 – 8,330,024 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 79.99 |
| Mean single sequence MFE | -24.43 |
| Consensus MFE | -12.18 |
| Energy contribution | -13.15 |
| Covariance contribution | 0.97 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.816947 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
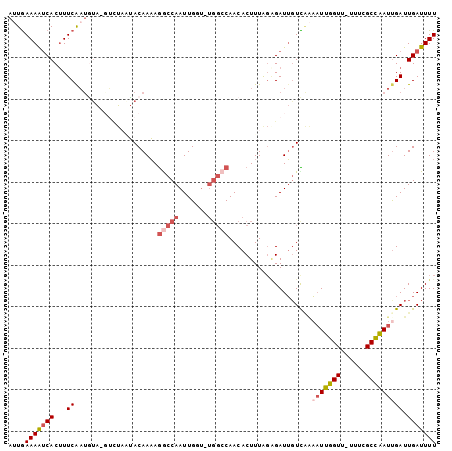
>2L_DroMel_CAF1 8329924 100 + 22407834 AUUGAAAAUCACUUUCAAUGUA-GUGUAAUACAAAAGGCCAAUUGGU-UGGCCAACACUUUAGUGAUUGUCAAAAUUGGUU-UUUCGCCAAUUGAUUGAUUUU .((((.(((((((.....((((-......))))...((((((....)-)))))........))))))).))))(((((((.-....))))))).......... ( -27.90) >DroSec_CAF1 8764 100 + 1 AUUGAAAAUCACUUUCAAUGUA-UUUUCGUACAAAAGGCCAAUUGGU-UGGCCAACACUUUAGAGAUUGUCAAAAUUGGUU-UUUCGCCAAUUGAUUGAUUUU ....(((((((.((((..((((-(....)))))...((((((....)-))))).........))))..((((..((((((.-....))))))))))))))))) ( -23.90) >DroSim_CAF1 9365 101 + 1 AUUGAAAAUCACUUUCAAUGUA-UUUUCGUACAAAAGGCCAAUUGGUUUGGCCAACACUUUAGAGAUUGUCAAAAUUGGUU-UUUCGCCAAUUGAUUGAUUUU ....(((((((.((((..((((-(....)))))...((((((.....)))))).........))))..((((..((((((.-....))))))))))))))))) ( -24.40) >DroEre_CAF1 9410 97 + 1 AUUGAAAAUCACUUUCGCUACAUGGCUAUUACAAAAGACCAAUUG------GCAACACUUUGGGGAUUGUUCCAAUGGGUCCGUUCGCCCAUUGAUUGAUUUU ....((((((((..(((((....)))............((((.((------....))..)))).))..))..((((((((......))))))))...)))))) ( -24.70) >DroYak_CAF1 9510 99 + 1 AUUGAAAAUCACUUUCAAUGUA-GCCUACUACAAAAGACCAAUUGCU-UGGCCAACACUUUAGAGAUUGUCAAAAUUGGUU--UUGGCCAAUCGAUUGAUUUU .((((((.....))))))((((-(....))))).((((.((((((.(-(((((((.(((.....((((.....))))))).--)))))))).)))))).)))) ( -26.20) >DroAna_CAF1 9163 99 + 1 AAUAAAAGCCACCUUCAACUUG-GUCCAAAACAAUAAGCCAAUUGAU-UGGCCAACACUUUACAGAUGUUUGGCUUUUGUU--UUUGCAAAUUGAUUGAUUUU (((((((((((((........)-))...(((((....(((((....)-)))).....((....)).)))))))))))))))--.................... ( -19.50) >consensus AUUGAAAAUCACUUUCAAUGUA_GUCUAAUACAAAAGGCCAAUUGGU_UGGCCAACACUUUAGAGAUUGUCAAAAUUGGUU_UUUCGCCAAUUGAUUGAUUUU ....(((((((...((....................(((((.......)))))....................(((((((......))))))))).))))))) (-12.18 = -13.15 + 0.97)


| Location | 8,329,924 – 8,330,024 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 79.99 |
| Mean single sequence MFE | -22.87 |
| Consensus MFE | -11.87 |
| Energy contribution | -11.98 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.816836 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
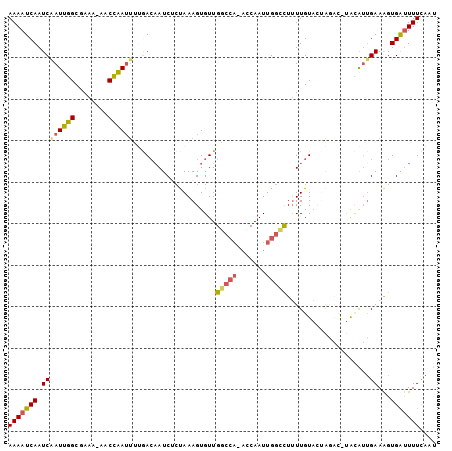
>2L_DroMel_CAF1 8329924 100 - 22407834 AAAAUCAAUCAAUUGGCGAAA-AACCAAUUUUGACAAUCACUAAAGUGUUGGCCA-ACCAAUUGGCCUUUUGUAUUACAC-UACAUUGAAAGUGAUUUUCAAU ..........((((((.....-..))))))((((.(((((((..(((((.(((((-(....))))))...((.....)).-.)))))...))))))).)))). ( -24.40) >DroSec_CAF1 8764 100 - 1 AAAAUCAAUCAAUUGGCGAAA-AACCAAUUUUGACAAUCUCUAAAGUGUUGGCCA-ACCAAUUGGCCUUUUGUACGAAAA-UACAUUGAAAGUGAUUUUCAAU ..........((((((.....-..))))))((((.((((.((..(((((.(((((-(....))))))((((....)))).-.)))))...)).)))).)))). ( -20.90) >DroSim_CAF1 9365 101 - 1 AAAAUCAAUCAAUUGGCGAAA-AACCAAUUUUGACAAUCUCUAAAGUGUUGGCCAAACCAAUUGGCCUUUUGUACGAAAA-UACAUUGAAAGUGAUUUUCAAU ..........((((((.....-..))))))((((.((((.((..(((((.((((((.....))))))((((....)))).-.)))))...)).)))).)))). ( -21.10) >DroEre_CAF1 9410 97 - 1 AAAAUCAAUCAAUGGGCGAACGGACCCAUUGGAACAAUCCCCAAAGUGUUGC------CAAUUGGUCUUUUGUAAUAGCCAUGUAGCGAAAGUGAUUUUCAAU ((((((.....(((((((((.(((((.(((((((((..........)))).)------)))).)))))))))).....))))...((....)))))))).... ( -24.80) >DroYak_CAF1 9510 99 - 1 AAAAUCAAUCGAUUGGCCAA--AACCAAUUUUGACAAUCUCUAAAGUGUUGGCCA-AGCAAUUGGUCUUUUGUAGUAGGC-UACAUUGAAAGUGAUUUUCAAU ..........((((((....--..))))))((((.((((.((........(((((-(....))))))...(((((....)-)))).....)).)))).)))). ( -24.10) >DroAna_CAF1 9163 99 - 1 AAAAUCAAUCAAUUUGCAAA--AACAAAAGCCAAACAUCUGUAAAGUGUUGGCCA-AUCAAUUGGCUUAUUGUUUUGGAC-CAAGUUGAAGGUGGCUUUUAUU (((..((.((((((((..((--(((((((((((((((.((....)))))(((...-.))).))))))).)))))))....-))))))))...))..))).... ( -21.90) >consensus AAAAUCAAUCAAUUGGCGAAA_AACCAAUUUUGACAAUCUCUAAAGUGUUGGCCA_ACCAAUUGGCCUUUUGUACUAGAC_UACAUUGAAAGUGAUUUUCAAU (((((((.((((((((........))))))....................(((((.......)))))....................))...))))))).... (-11.87 = -11.98 + 0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:39:05 2006