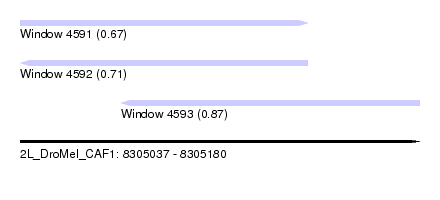
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,305,037 – 8,305,180 |
| Length | 143 |
| Max. P | 0.865254 |

| Location | 8,305,037 – 8,305,140 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 82.43 |
| Mean single sequence MFE | -30.89 |
| Consensus MFE | -21.43 |
| Energy contribution | -22.07 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.665994 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8305037 103 + 22407834 UUUAUCUAUUGUAGUCCCAACAACAGCUUUUACCAGUUGGACUGCGGAAACUCCAGCAUUUAUGCUCGUCCCAAGCCACCGCCAUAUAAGGGCGGUACUUUGG ..........(((((((.(((..............))))))))))((.....))((((....))))....(((((..((((((.......))))))..))))) ( -30.04) >DroVir_CAF1 1512 99 + 1 ----UCAAUUGCAGUCCAAGCAAUAGCUUGUAUCAGCUGGACACUGGCGCCUCCGGCAUCUAUGCACGGCCCAAGCCGCCACCCUAUAAGGGCGGCACUUUGG ----.....((((...(((((....))))).....((((((..(....)..)))))).....)))).(((....)))(((.(((.....))).)))....... ( -33.70) >DroSec_CAF1 1503 103 + 1 UUUAUCUAUUGCAGUCCCAACAACAGCUUUUACCAGUUGGACUGCGGAAACUCCAGCAUUUAUGCUCGUCCCAAGCCACCACCAUAUAAGGGCGGCACUUUGG ..........(((((((.(((..............))))))))))((.....))((((....))))....(((((((.((..........)).)))...)))) ( -25.44) >DroEre_CAF1 1524 103 + 1 UUUGUCUAUUGCAGUCCCAACAACAGCUUUUACCAGUUGGACUGCGGAAACUCCAGCAUUUAUGCACGACCCAAGCCACCGCCCUAUAAGGGCGGCACUUUGG ...(((....(((((((.(((..............))))))))))((.....)).(((....)))..)))(((((...((((((.....))))))...))))) ( -35.34) >DroYak_CAF1 1478 103 + 1 UUUGUUUAUUGCAGUCCCAACAACAGCCUUUACCAGUUGGACUGCGGAAACUCCAGCAUCUAUGCCCGCCCCAAGCCGCCGCCGUACAAGGGCGGCACUUUGG ...((((.(((((((((.(((..............))))))))))))))))....(((....))).....(((((..((((((.......))))))..))))) ( -33.94) >DroMoj_CAF1 1487 100 + 1 C---UUAUUUGCAGUCCAAACAAUAGUUUAUAUCAGCUGGAUACAGGUACCUCGGGCAUAUACGCCCGCCCUAAGCCUCCGCCGUAUAAGGGCGGCACUUUGG .---.((((((..(((((....(((.....)))....))))).))))))...(((((......))))).((.(((...(((((.......)))))..))).)) ( -26.90) >consensus UUU_UCUAUUGCAGUCCCAACAACAGCUUUUACCAGUUGGACUGCGGAAACUCCAGCAUUUAUGCACGCCCCAAGCCACCGCCAUAUAAGGGCGGCACUUUGG ........(((((((((.(((..............))))))))))))........((......)).....(((((...(((((.......)))))...))))) (-21.43 = -22.07 + 0.64)

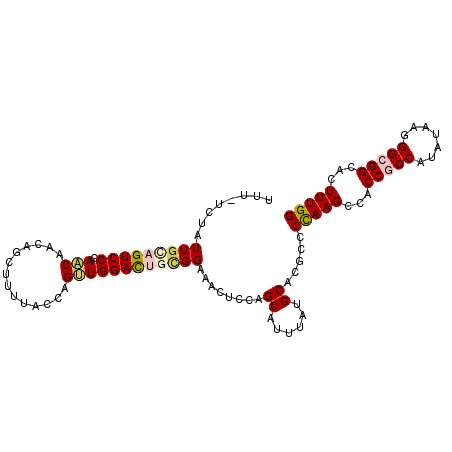
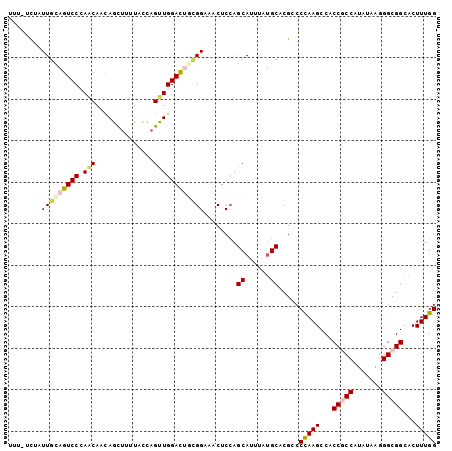
| Location | 8,305,037 – 8,305,140 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 82.43 |
| Mean single sequence MFE | -37.05 |
| Consensus MFE | -24.51 |
| Energy contribution | -24.32 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.711547 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8305037 103 - 22407834 CCAAAGUACCGCCCUUAUAUGGCGGUGGCUUGGGACGAGCAUAAAUGCUGGAGUUUCCGCAGUCCAACUGGUAAAAGCUGUUGUUGGGACUACAAUAGAUAAA (((((((((((((.......)))))).)))).((((.((((....))))((.....))...))))...)))......(((((((.......)))))))..... ( -31.70) >DroVir_CAF1 1512 99 - 1 CCAAAGUGCCGCCCUUAUAGGGUGGCGGCUUGGGCCGUGCAUAGAUGCCGGAGGCGCCAGUGUCCAGCUGAUACAAGCUAUUGCUUGGACUGCAAUUGA---- ((((..((((((((.....))))))))..))))....((((.....((.(((.((....)).))).)).....(((((....)))))...)))).....---- ( -40.20) >DroSec_CAF1 1503 103 - 1 CCAAAGUGCCGCCCUUAUAUGGUGGUGGCUUGGGACGAGCAUAAAUGCUGGAGUUUCCGCAGUCCAACUGGUAAAAGCUGUUGUUGGGACUGCAAUAGAUAAA ((((...((((((((.....)).))))))))))..(.((((....)))).).......((((((((((.(((....)))...))).))))))).......... ( -35.20) >DroEre_CAF1 1524 103 - 1 CCAAAGUGCCGCCCUUAUAGGGCGGUGGCUUGGGUCGUGCAUAAAUGCUGGAGUUUCCGCAGUCCAACUGGUAAAAGCUGUUGUUGGGACUGCAAUAGACAAA ((((..((((((((.....))))))))..))))(((..(((....))).((.....))((((((((((.(((....)))...))).)))))))....)))... ( -41.50) >DroYak_CAF1 1478 103 - 1 CCAAAGUGCCGCCCUUGUACGGCGGCGGCUUGGGGCGGGCAUAGAUGCUGGAGUUUCCGCAGUCCAACUGGUAAAGGCUGUUGUUGGGACUGCAAUAAACAAA ((((..(((((((.......)))))))..))))..(.((((....)))).).((((..((((((((((.(((....)))...))).)))))))...))))... ( -39.20) >DroMoj_CAF1 1487 100 - 1 CCAAAGUGCCGCCCUUAUACGGCGGAGGCUUAGGGCGGGCGUAUAUGCCCGAGGUACCUGUAUCCAGCUGAUAUAAACUAUUGUUUGGACUGCAAAUAA---G .....((((((((((.....(((....))).)))))(((((....)))))..))))).(((((((((..((((.....))))..))))).)))).....---. ( -34.50) >consensus CCAAAGUGCCGCCCUUAUACGGCGGUGGCUUGGGACGAGCAUAAAUGCUGGAGUUUCCGCAGUCCAACUGGUAAAAGCUGUUGUUGGGACUGCAAUAGA_AAA ((.((((((((((.......)))))).)))).)).(.((((....)))).).........((((((((..............))).)))))............ (-24.51 = -24.32 + -0.19)

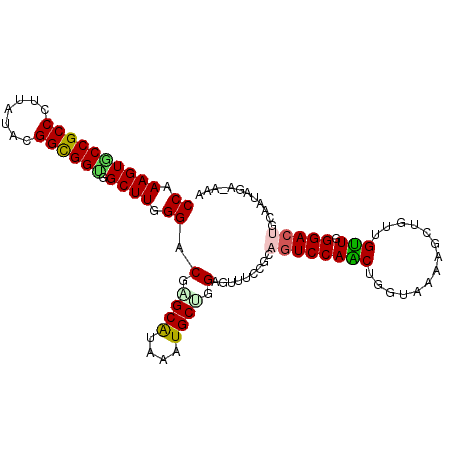
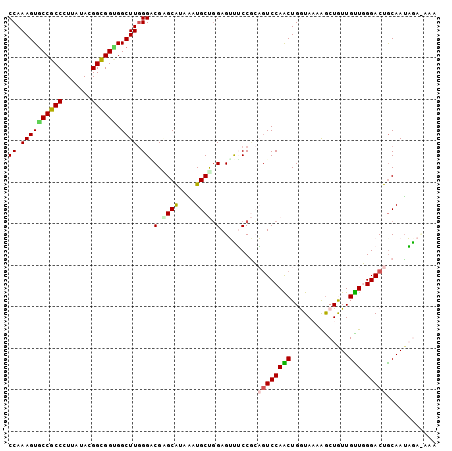
| Location | 8,305,073 – 8,305,180 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 80.31 |
| Mean single sequence MFE | -42.78 |
| Consensus MFE | -27.23 |
| Energy contribution | -27.12 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.865254 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8305073 107 - 22407834 ACUCGGGGAGAUGUUUGUCAAAUGUCCACUCCAUUUCGACCCAAAGUACCGCCCUUAUAUGGCGGUGGCUUGGGACGAGCAUAAAUGCUGGAGUUUCCGCAGUCCAA (((((((((.((.........)).)))((((((..(((.(((((..(((((((.......)))))))..))))).)))(((....)))))))))..))).))).... ( -40.80) >DroVir_CAF1 1544 107 - 1 ACUCGCGGCGAUGUUUGUCAAAUGUCCACGCCAUUGCGUCCCAAAGUGCCGCCCUUAUAGGGUGGCGGCUUGGGCCGUGCAUAGAUGCCGGAGGCGCCAGUGUCCAG ...((((((((((((.....))))))...(((.......(((((..((((((((.....))))))))..)))))(((.(((....)))))).)))))).)))..... ( -44.50) >DroGri_CAF1 1834 107 - 1 ACUCGAGGCGAUGUCUGUCAAAUAUCCACACCAUUCCGCCCGAGAGUGCCGCCCUUAUACGGUGGCGGUUUGGGACGUGCAUAUAAACCAGAGGCGCCACCAUCCAU ......((((.(.((((....((((.(((.(((..((((((....).((((........)))))))))..)))...))).))))....)))).)))))......... ( -36.40) >DroYak_CAF1 1514 107 - 1 ACUCGGGGAGAUGUUUGUCAAAUGUCCACUCCAUUUCGUCCCAAAGUGCCGCCCUUGUACGGCGGCGGCUUGGGGCGGGCAUAGAUGCUGGAGUUUCCGCAGUCCAA (((((((((.((.........)).)))(((((....((((((((..(((((((.......)))))))..))))))))((((....)))))))))..))).))).... ( -46.50) >DroMoj_CAF1 1520 107 - 1 ACUCGCGGCGAUGUUUGUCAAAUGUCCACGCCAUUGCGACCCAAAGUGCCGCCCUUAUACGGCGGAGGCUUAGGGCGGGCGUAUAUGCCCGAGGUACCUGUAUCCAG .((((.((((((....)))..((((((.(((....))).(((.((((.(((((.......)))))..)))).))).))))))....))))))).............. ( -41.20) >DroAna_CAF1 1538 98 - 1 ACUCGCGGGGAUGUUUGUCAAAUAUCCACUCCGUUCCGGCCCAGAGUGCCGCCCUUGUACGGAGGGGGCUUGGGCCGGGCAUAAAUGCUGGAG---------UCCAA ......(((((((((.....)))))))(((((...(((((((((...(((.(((((.....)))))))))))))))))(((....))).))))---------))).. ( -47.30) >consensus ACUCGCGGCGAUGUUUGUCAAAUGUCCACUCCAUUCCGACCCAAAGUGCCGCCCUUAUACGGCGGCGGCUUGGGACGGGCAUAAAUGCCGGAGGUGCCACAGUCCAA .(((..((((.((..(((...)))..))))))..((((.(((((..(((((((.......)))))))..))))).))))...........))).............. (-27.23 = -27.12 + -0.11)

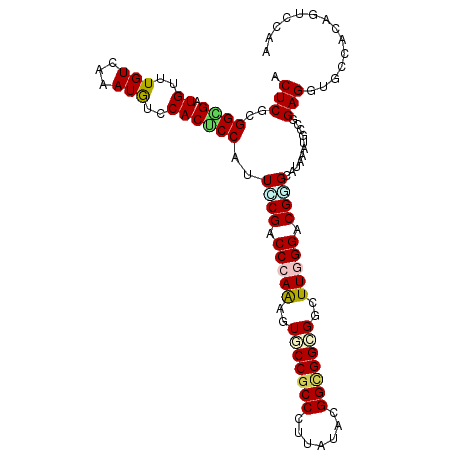
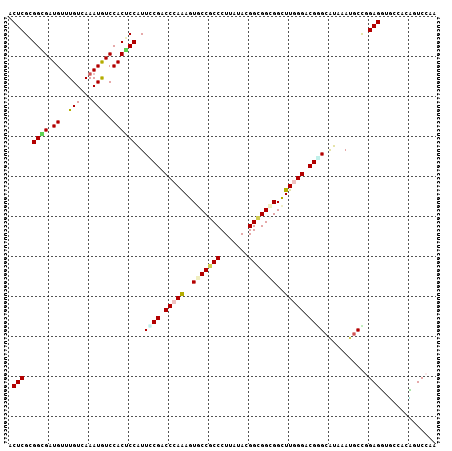
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:38:51 2006