
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,291,781 – 8,291,915 |
| Length | 134 |
| Max. P | 0.746598 |

| Location | 8,291,781 – 8,291,875 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 78.79 |
| Mean single sequence MFE | -16.70 |
| Consensus MFE | -10.77 |
| Energy contribution | -11.28 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.605498 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8291781 94 - 22407834 AAUAGCUUUGUGCCAUUUUGGCAAAGUAGGAAAGCUAUUAAAUGUACUUAUUAACUUACUUUCAACACAAAAAUCGAUUGCUAUCUUAGGCAA------U (((((((((.((((.....)))).......))))))))).....................................((((((......)))))------) ( -16.90) >DroSim_CAF1 72784 85 - 1 AAUAGCUUUGUGCCAUUUUGGCAAAGUAGGAAAGCUAUUAAAUGUACUUAUUAACUUA---------CAAAAAUCGAUUGCUAUCUUAGGCAA------C (((((((((.((((.....)))).......)))))))))...................---------..........(((((......)))))------. ( -16.20) >DroEre_CAF1 72396 93 - 1 AAUAGCUUUGUGCCAUUUUGGCAAAGUAGGAAAGCUAUUAAAAGUAG-UA-UAACUUACUAUUAACA-AAAAAUAA----CUUACUUACAAAAAAUCGAU (((((((((.((((.....)))).......)))))))))...(((((-((-.....)))))))....-........----.................... ( -17.20) >DroYak_CAF1 75153 92 - 1 AAUAGCUUUGUGCCAUUUUGGCAAACUAGGCACGCUAUUAAAUGUACUUA-CAACUUACUAUUAACA-AAAAAUUGGUUGCUAACUUACAUAA------U ((((((...(((((..............)))))))))))..(((((.(((-(((((...........-.......))))).)))..)))))..------. ( -16.51) >consensus AAUAGCUUUGUGCCAUUUUGGCAAAGUAGGAAAGCUAUUAAAUGUACUUA_UAACUUACUAUUAACA_AAAAAUCGAUUGCUAACUUACACAA______U (((((((((.((((.....)))).......)))))))))............................................................. (-10.77 = -11.28 + 0.50)

| Location | 8,291,810 – 8,291,915 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 91.57 |
| Mean single sequence MFE | -21.82 |
| Consensus MFE | -18.26 |
| Energy contribution | -18.82 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.746598 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
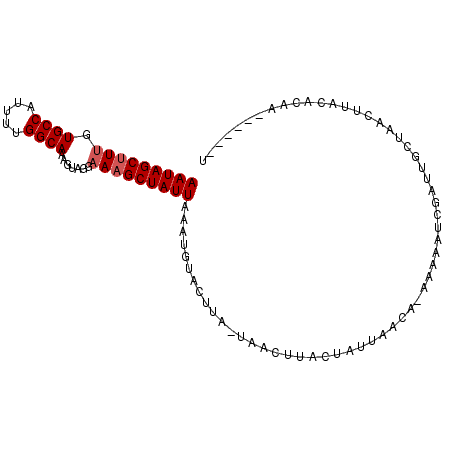
>2L_DroMel_CAF1 8291810 105 + 22407834 UUGAAAGUAAGUUAAUAAGUACAUUUAAUAGCUUUCCUACUUUGCCAAAAUGGCACAAAGCUAUUGAAAACGUAAGCUCCUUGUGUUAAUGAUUUAUGCCGAGUC (((...((((((((.(((.((((((((((((((((.......((((.....)))).))))))))))))..(....).....))))))).))))))))..)))... ( -22.40) >DroSim_CAF1 72811 98 + 1 -------UAAGUUAAUAAGUACAUUUAAUAGCUUUCCUACUUUGCCAAAAUGGCACAAAGCUAUUGAAAACGUAAGCUCCUUGUGUUAAUGAUUUAUGCCAAGUC -------(((((((.(((.((((((((((((((((.......((((.....)))).))))))))))))..(....).....))))))).)))))))......... ( -20.00) >DroEre_CAF1 72426 103 + 1 UUAAUAGUAAGUUA-UA-CUACUUUUAAUAGCUUUCCUACUUUGCCAAAAUGGCACAAAGCUAUUGAAAACGUAAGCUCCUUGUGUUAAUGAUUUAUGCCAAGUC ......((((((((-(.-.....((((((((((((.......((((.....)))).))))))))))))((((((((...)))))))).)))))))))........ ( -22.10) >DroYak_CAF1 75181 104 + 1 UUAAUAGUAAGUUG-UAAGUACAUUUAAUAGCGUGCCUAGUUUGCCAAAAUGGCACAAAGCUAUUGAAAACGUAAGCUCCUUGUGUUAAUGAUUUAUGCCAAGUC ......((((((..-(.((((((((((((((((((((..((((....)))))))))...)))))))))..(....).....)))))).)..))))))........ ( -22.80) >consensus UUAAUAGUAAGUUA_UAAGUACAUUUAAUAGCUUUCCUACUUUGCCAAAAUGGCACAAAGCUAUUGAAAACGUAAGCUCCUUGUGUUAAUGAUUUAUGCCAAGUC ......((((((((.........((((((((((((.......((((.....)))).))))))))))))((((((((...))))))))..))))))))........ (-18.26 = -18.82 + 0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:38:48 2006