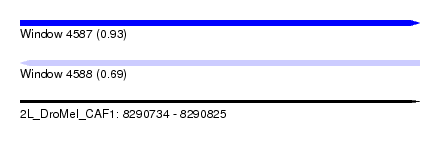
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,290,734 – 8,290,825 |
| Length | 91 |
| Max. P | 0.928228 |

| Location | 8,290,734 – 8,290,825 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.48 |
| Mean single sequence MFE | -26.12 |
| Consensus MFE | -20.48 |
| Energy contribution | -20.98 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928228 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
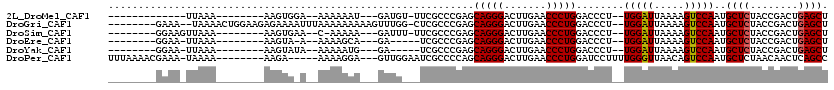
>2L_DroMel_CAF1 8290734 91 + 22407834 AGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCA--AGGGUCCAGGGUUCAAGUCCCUGCUCGGGCGAA-ACAUC---AUUUUUU--UCCACUU--------UUUAA------------- .(((((((.((..((((.((((((((((.....)--))))))))).)))).....)).))).))))...-.....---.......--.......--------.....------------- ( -24.10) >DroGri_CAF1 89292 107 + 1 AGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCA--AGGGUCCAGGGUUCAAGUCCCUGCUCGGGCGAG-CCAAACUUUUUUUUUAAAUUUUCUCUUCCAGUUUUA--UUUC-------- .((((.(((....(((((.(((((((((.....)--))))))))(((.......)))))))).))))))-)...................................--....-------- ( -27.80) >DroSim_CAF1 71767 95 + 1 AGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCA--AGGGUCCAGGGUUCAAGUCCCUGCUCGGGCGAA-AAAUC---UUUUU-G--UUCACUU--------UUUAACUUCC-------- .....(((..(..(((((.(((((((((.....)--))))))))(((.......))))))))(((((((-(....---.))))-)--)))....--------.)..)))...-------- ( -26.50) >DroEre_CAF1 71215 90 + 1 AGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCA--AGGGUCCAGGGUUCAAGUCCCUGCUCGGGCGA-----UC---UGCUUUU--U-UACUU--------UUUAA-UUCC-------- (((...((((.(.(((((.(((((((((.....)--))))))))(((.......)))))))).).)))-----).---.)))...--.-.....--------.....-....-------- ( -27.40) >DroYak_CAF1 73925 91 + 1 AGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCA--AGGGUCCAGGGUUCAAGUCCCUGCUCGGGCGA-----UC---CAUUUUU--UAUACUU--------UUUAA-UUCC-------- ......((((.(.(((((.(((((((((.....)--))))))))(((.......)))))))).).)))-----).---.......--.......--------.....-....-------- ( -24.50) >DroPer_CAF1 91904 103 + 1 GGCUGAGUUGUUAGAGCAUUGGACUGUUAACCCAAAAGGAUCCAGGGUUCAAGUCCCUGCUGGGGCGAUUCCAAC---UCCUUUU-----UCUU--------UUUUA-UUUCGUUUUAAA ....((((((((..((((..(((((...(((((...........)))))..))))).))))..))))))))....---.......-----....--------.....-............ ( -26.40) >consensus AGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCA__AGGGUCCAGGGUUCAAGUCCCUGCUCGGGCGA___CAUC___UUUUUUU__U_UACUU________UUUAA_UUCC________ .((((........)))).(((((((((..((((....))))..)))))))))((((......))))...................................................... (-20.48 = -20.98 + 0.50)

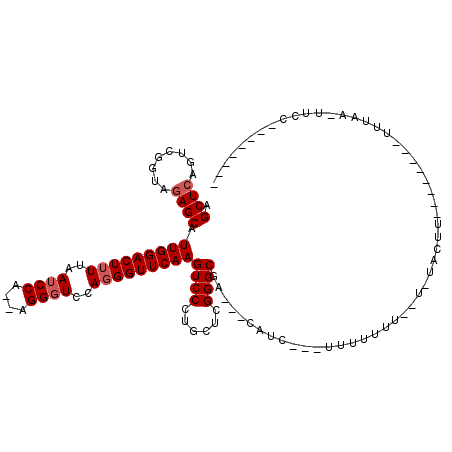
| Location | 8,290,734 – 8,290,825 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.48 |
| Mean single sequence MFE | -23.12 |
| Consensus MFE | -16.10 |
| Energy contribution | -16.13 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.686779 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8290734 91 - 22407834 -------------UUAAA--------AAGUGGA--AAAAAAU---GAUGU-UUCGCCCGAGCAGGGACUUGAACCCUGGACCCU--UGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCU -------------.....--------...((((--.......---((...-.))..((((((((((.......)))))....))--)))........))))..((((........)))). ( -21.20) >DroGri_CAF1 89292 107 - 1 --------GAAA--UAAAACUGGAAGAGAAAAUUUAAAAAAAAAGUUUGG-CUCGCCCGAGCAGGGACUUGAACCCUGGACCCU--UGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCU --------....--......((((.(((.((((((.......))))))..-)))..((((((((((.......)))))....))--)))........))))..((((........)))). ( -26.00) >DroSim_CAF1 71767 95 - 1 --------GGAAGUUAAA--------AAGUGAA--C-AAAAA---GAUUU-UUCGCCCGAGCAGGGACUUGAACCCUGGACCCU--UGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCU --------(((..((((.--------..(((((--.-((...---..)).-)))))((((((((((.......)))))....))--))).))))...)))...((((........)))). ( -24.50) >DroEre_CAF1 71215 90 - 1 --------GGAA-UUAAA--------AAGUA-A--AAAAGCA---GA-----UCGCCCGAGCAGGGACUUGAACCCUGGACCCU--UGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCU --------....-.....--------.....-.--.....((---(.-----(((...((((((((.((........)).)))(--(((((.....)))))))))))...)))))).... ( -22.50) >DroYak_CAF1 73925 91 - 1 --------GGAA-UUAAA--------AAGUAUA--AAAAAUG---GA-----UCGCCCGAGCAGGGACUUGAACCCUGGACCCU--UGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCU --------((..-.....--------.......--.....((---(.-----....)))..(((((.......)))))...))(--(((((.....)))))).((((........)))). ( -20.80) >DroPer_CAF1 91904 103 - 1 UUUAAAACGAAA-UAAAA--------AAGA-----AAAAGGA---GUUGGAAUCGCCCCAGCAGGGACUUGAACCCUGGAUCCUUUUGGGUUAACAGUCCAAUGCUCUAACAACUCAGCC ............-.....--------....-----.....((---(((((........)((((.(((((..(((((...........)))))...)))))..))))....)))))).... ( -23.70) >consensus ________GGAA_UUAAA________AAGUA_A__AAAAAAA___GAUG___UCGCCCGAGCAGGGACUUGAACCCUGGACCCU__UGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCU .............................................................(((((.......)))))........(((((.....)))))..((((........)))). (-16.10 = -16.13 + 0.03)
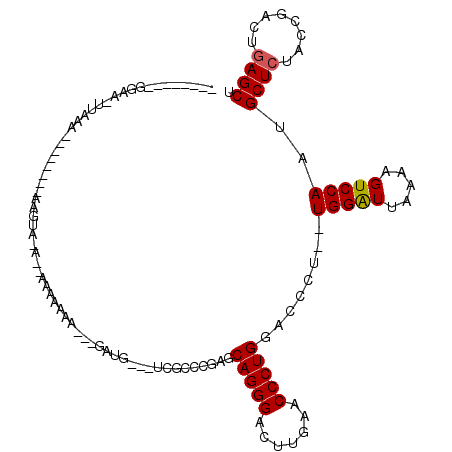
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:38:47 2006