
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,272,577 – 8,272,717 |
| Length | 140 |
| Max. P | 0.841263 |

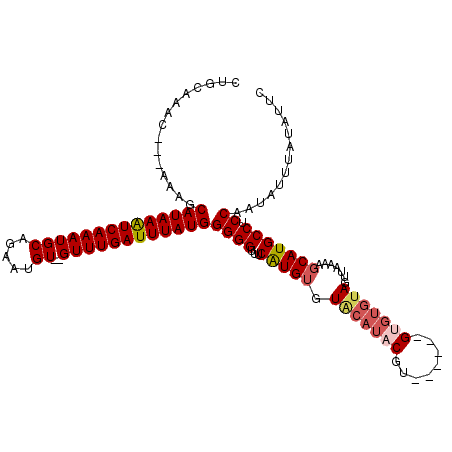
| Location | 8,272,577 – 8,272,687 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 87.04 |
| Mean single sequence MFE | -27.72 |
| Consensus MFE | -18.86 |
| Energy contribution | -19.92 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.830952 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8272577 110 - 22407834 CUGCAAAC---AAAGCAUAAAUCAAAUGCAGAAUGU--GUUUGAUUUAUGGGGGUUUCAUGUGUGCAUACAUAUGUACGUGUGUAGUUAAAAGCAUGCCACCAAUAUUUAUACUC ........---....((((((((((((((.....))--))))))))))))..(((..(((((..((.(((((((....))))))))).....)))))..)))............. ( -30.60) >DroSec_CAF1 50551 109 - 1 CUGCAAACAACAAAGCAUAAAUCAAAUGCAGAAUGUGUGUUUGAUUUAUGGGGGUUUCAUGUGUACAUACGU------GUGUGUACUUAAAAGCAUGCCUCCAAUAUUUAUAUUC .(((..........)))(((((((((((((.....)))))))))))))((((((...((((((((((((...------.)))))))......)))))))))))............ ( -30.60) >DroSim_CAF1 51440 109 - 1 CUGCAAACAACAAAGCAUAAAUCAAAUGCAGAAUGUGUGUUUGAUUUAUGGGGGUUUCGUGUGUACAUACGU------GUGUGUAGUUAAAAGCAUGCCUCCAAUAUUUAUAUUC ...............(((((((((((((((.....)))))))))))))))((((...(((((....))))).------((((((........))))))))))............. ( -27.60) >DroEre_CAF1 52922 99 - 1 CUGCAAAC---AAAGCAUAAGUCAAAUGCAGAAUGU--GUUUGAUUUAUGGGGGUUUCAUGUGUAC--ACGA------G---GUAGUUAAAAUCAUGCCUCCAAUAUUUGUAUUC .((((((.---....((((((((((((((.....))--))))))))))))((((...((((.....--((..------.---...))......)))).))))....))))))... ( -25.40) >DroYak_CAF1 54397 101 - 1 CUGCACAC---AAAGCAUAAAUCAAAUGCAUAAUGU--GUUUGAUUUAUGGGGGUUUCAUGUGUUCGUACGA------G---UUAGUUAAAAGCAUGCCACCAAUAUUUAUAUUC ........---....((((((((((((((.....))--))))))))))))..(((..(((((......((..------.---...)).....)))))..)))............. ( -24.40) >consensus CUGCAAAC___AAAGCAUAAAUCAAAUGCAGAAUGU__GUUUGAUUUAUGGGGGUUUCAUGUGUACAUACGU______GUGUGUAGUUAAAAGCAUGCCUCCAAUAUUUAUAUUC ...............((((((((((((((.....))..))))))))))))((((...(((((.(((((((........))))))).......))))))).))............. (-18.86 = -19.92 + 1.06)


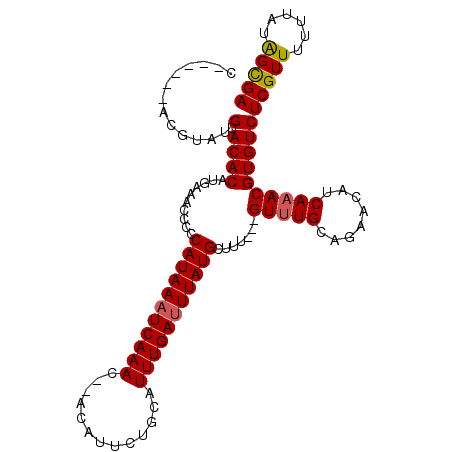
| Location | 8,272,613 – 8,272,717 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 89.31 |
| Mean single sequence MFE | -19.90 |
| Consensus MFE | -15.26 |
| Energy contribution | -15.34 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.688216 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8272613 104 + 22407834 CGUACAUAUGUAUGCACACAUGAAACCCCCAUAAAUCAAAC--ACAUUCUGCAUUUGAUUUAUGCUUU---GUUUGCAGAACAUCAAACGUGUCUCGUUU-UUAUGGCGA (((.((((...(((.((((..........(((((((((((.--.(.....)..)))))))))))....---(((((........)))))))))..)))..-.))))))). ( -21.60) >DroSec_CAF1 50587 104 + 1 C------ACGUAUGUACACAUGAAACCCCCAUAAAUCAAACACACAUUCUGCAUUUGAUUUAUGCUUUGUUGUUUGCAGAACAUCAAACGUGUCUCGUUUUUUAUAGCGA (------((((((((..............(((((((((((..((.....))..))))))))))).(((((.....)))))))))...)))))..(((((......))))) ( -21.10) >DroSim_CAF1 51476 104 + 1 C------ACGUAUGUACACACGAAACCCCCAUAAAUCAAACACACAUUCUGCAUUUGAUUUAUGCUUUGUUGUUUGCAGAACAUCAAACGUGUCUCGUUUUUUAUAGCGA (------((((.((((.(((((((.....(((((((((((..((.....))..))))))))))).)))).))).))))(.....)..)))))..(((((......))))) ( -21.60) >DroEre_CAF1 52955 96 + 1 C------UCGU--GUACACAUGAAACCCCCAUAAAUCAAAC--ACAUUCUGCAUUUGACUUAUGCUUU---GUUUGCAGAACAUCAAACGUGUCUCGUUU-UUAUAGCGA .------((((--(....)))))................((--((.....((((.......))))...---(((((........))))))))).(((((.-....))))) ( -15.90) >DroYak_CAF1 54430 99 + 1 C------UCGUACGAACACAUGAAACCCCCAUAAAUCAAAC--ACAUUAUGCAUUUGAUUUAUGCUUU---GUGUGCAGAACAUCAAACGUGUCUCGUUUUUUAUAGUGA .------....((((((((..........(((((((((((.--.(.....)..))))))))))).(((---((((.....))).)))).)))).))))............ ( -19.30) >consensus C______ACGUAUGUACACAUGAAACCCCCAUAAAUCAAAC__ACAUUCUGCAUUUGAUUUAUGCUUU___GUUUGCAGAACAUCAAACGUGUCUCGUUUUUUAUAGCGA .............(.((((..........(((((((((((.............))))))))))).......(((((........))))))))))(((((......))))) (-15.26 = -15.34 + 0.08)


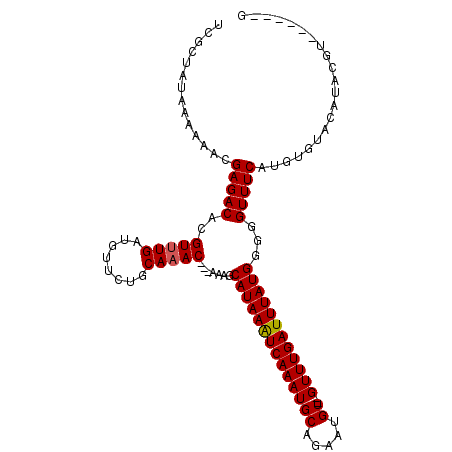
| Location | 8,272,613 – 8,272,717 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 89.31 |
| Mean single sequence MFE | -27.34 |
| Consensus MFE | -19.48 |
| Energy contribution | -19.52 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.841263 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8272613 104 - 22407834 UCGCCAUAA-AAACGAGACACGUUUGAUGUUCUGCAAAC---AAAGCAUAAAUCAAAUGCAGAAUGU--GUUUGAUUUAUGGGGGUUUCAUGUGUGCAUACAUAUGUACG .........-....(((((..(((((........)))))---....((((((((((((((.....))--))))))))))))...)))))....((((((....)))))). ( -29.90) >DroSec_CAF1 50587 104 - 1 UCGCUAUAAAAAACGAGACACGUUUGAUGUUCUGCAAACAACAAAGCAUAAAUCAAAUGCAGAAUGUGUGUUUGAUUUAUGGGGGUUUCAUGUGUACAUACGU------G ..............(((((..(((((........))))).......(((((((((((((((.....)))))))))))))))...)))))(((((....)))))------. ( -26.10) >DroSim_CAF1 51476 104 - 1 UCGCUAUAAAAAACGAGACACGUUUGAUGUUCUGCAAACAACAAAGCAUAAAUCAAAUGCAGAAUGUGUGUUUGAUUUAUGGGGGUUUCGUGUGUACAUACGU------G ..(.((((....(((((((..(((((........))))).......(((((((((((((((.....)))))))))))))))...))))))).)))))......------. ( -29.10) >DroEre_CAF1 52955 96 - 1 UCGCUAUAA-AAACGAGACACGUUUGAUGUUCUGCAAAC---AAAGCAUAAGUCAAAUGCAGAAUGU--GUUUGAUUUAUGGGGGUUUCAUGUGUAC--ACGA------G (((.((((.-....(((((..(((((........)))))---....((((((((((((((.....))--))))))))))))...)))))...)))).--.)))------. ( -25.30) >DroYak_CAF1 54430 99 - 1 UCACUAUAAAAAACGAGACACGUUUGAUGUUCUGCACAC---AAAGCAUAAAUCAAAUGCAUAAUGU--GUUUGAUUUAUGGGGGUUUCAUGUGUUCGUACGA------G ............(((((.(((((..(((.(((.((....---...))(((((((((((((.....))--)))))))))))))).)))..))))))))))....------. ( -26.30) >consensus UCGCUAUAAAAAACGAGACACGUUUGAUGUUCUGCAAAC___AAAGCAUAAAUCAAAUGCAGAAUGU__GUUUGAUUUAUGGGGGUUUCAUGUGUACAUACGU______G ..............(((((..(((((........))))).......((((((((((((((.....))..))))))))))))...)))))..................... (-19.48 = -19.52 + 0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:38:40 2006