
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
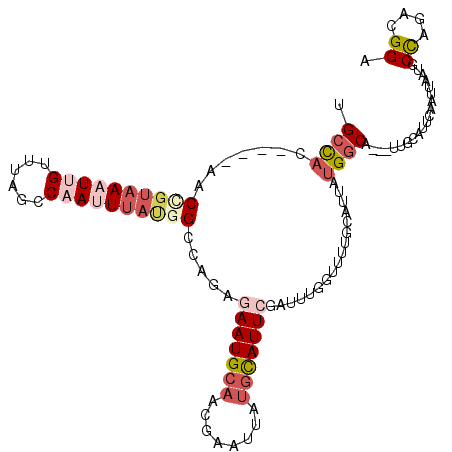
| Location | 8,268,129 – 8,268,258 |
| Length | 129 |
| Max. P | 0.953169 |

| Location | 8,268,129 – 8,268,237 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 80.18 |
| Mean single sequence MFE | -29.67 |
| Consensus MFE | -13.50 |
| Energy contribution | -15.37 |
| Covariance contribution | 1.86 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.747757 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8268129 108 + 22407834 CGCCAC----UACCGUAAAUUGUUUAGCCAAUUUAUGGCCAGAGAAUGCAUCGAAUUAUGCAUUCGAUUUGGUUUUACACUAUGGCA---UUGCAUUCAAUUAAUGCCAGACGGA ..((..----..((((((((((......))))))))))(((((((((((((......))))))))..)))))..........(((((---(((........))))))))...)). ( -33.00) >DroSec_CAF1 46174 104 + 1 UGCCAC----AAC----AAUUGGCUAAACAAUUUACGGCCAGAGAAUGCAACAAAUUAUGCAUUCGAUUUGGUUUUGCAUUAUGGCA---UUGCAUUCAAUUAAUGCCAGACGGA .((((.----...----...))))...........(((((((((((((((........)))))))..)))))).........(((((---(((........))))))))..)).. ( -28.00) >DroSim_CAF1 46261 108 + 1 UGCCAC----AACCGUAAUUUGUUUAGCCAAUUUACGGCCAGAGAAUGCAACAAAUUAUGCAUUCGAUUUGGUUUUGCAUUAUGGCA---UUGCAUUCAAUUAAUGCCAGACGGA (((...----..((((((.(((......))).))))))((((((((((((........)))))))..)))))....)))...(((((---(((........))))))))...... ( -30.80) >DroEre_CAF1 48464 103 + 1 UGCCAC----AACUGUAAAUGGCUUAGCCAAUUUAUGGCAAGAAAAUGCAUCGCAUUAUGCAUUCGAUUUGGUUUUGCAUUCUGGC--------AUUCAAUUAAUGCCAGACGGA ..((..----...........((..((((((......(((....(((((...))))).))).......))))))..))..((((((--------(((.....))))))))).)). ( -28.62) >DroYak_CAF1 49803 103 + 1 UGCCAC----AACUGUAAAUGGUUUAGCCAAUUUAUGGCAAGAAAAUGCAUCGAAUUAUGCAUUCGAUUUGGUUUUGCAUUCUGGC--------AUUCAAUUAAUGCCAGACGGA ..((..----...........((..((((((......(((......)))(((((((.....)))))))))))))..))..((((((--------(((.....))))))))).)). ( -29.00) >DroAna_CAF1 54420 110 + 1 UGCUUUGACGAACGGUAAUUUGCUUAGCCAAUUUAUGGCCAG-GAAUGCAACGAAUUAAAUAUUCGAUACUGUCUAGAAACCUGCCACCCUGGCAUUCA----AUGCUAGACGGA ........((((...((((((((((.((((.....)))).))-).......)))))))....))))...((((((((.....((((.....))))....----...)))))))). ( -28.61) >consensus UGCCAC____AACCGUAAAUUGUUUAGCCAAUUUAUGGCCAGAGAAUGCAACGAAUUAUGCAUUCGAUUUGGUUUUGCAUUAUGGCA___UUGCAUUCAAUUAAUGCCAGACGGA .((((.......((((((((((......)))))))))).....(((((((........))))))).................))))....................((....)). (-13.50 = -15.37 + 1.86)


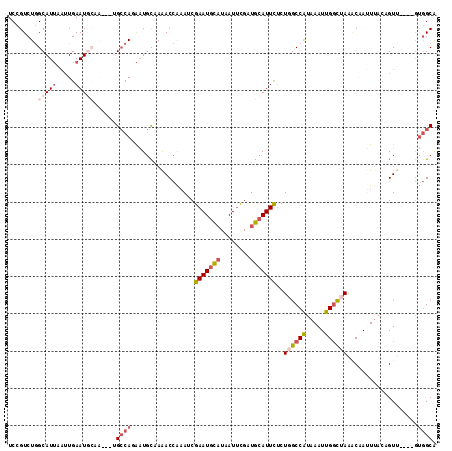
| Location | 8,268,129 – 8,268,237 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 80.18 |
| Mean single sequence MFE | -31.10 |
| Consensus MFE | -15.69 |
| Energy contribution | -17.13 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.18 |
| Mean z-score | -3.22 |
| Structure conservation index | 0.50 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.953169 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8268129 108 - 22407834 UCCGUCUGGCAUUAAUUGAAUGCAA---UGCCAUAGUGUAAAACCAAAUCGAAUGCAUAAUUCGAUGCAUUCUCUGGCCAUAAAUUGGCUAAACAAUUUACGGUA----GUGGCG ........(((((.....)))))..---.((((((.((((((........((((((((......))))))))..((((((.....)))))).....)))))).).----))))). ( -34.00) >DroSec_CAF1 46174 104 - 1 UCCGUCUGGCAUUAAUUGAAUGCAA---UGCCAUAAUGCAAAACCAAAUCGAAUGCAUAAUUUGUUGCAUUCUCUGGCCGUAAAUUGUUUAGCCAAUU----GUU----GUGGCA ........(((((.....)))))..---(((((((((.............(((((((........)))))))..((((.............))))...----)))----)))))) ( -27.82) >DroSim_CAF1 46261 108 - 1 UCCGUCUGGCAUUAAUUGAAUGCAA---UGCCAUAAUGCAAAACCAAAUCGAAUGCAUAAUUUGUUGCAUUCUCUGGCCGUAAAUUGGCUAAACAAAUUACGGUU----GUGGCA ........(((((.....)))))..---((((((((.......(((....(((((((........)))))))..)))((((((.(((......))).))))))))----)))))) ( -33.20) >DroEre_CAF1 48464 103 - 1 UCCGUCUGGCAUUAAUUGAAU--------GCCAGAAUGCAAAACCAAAUCGAAUGCAUAAUGCGAUGCAUUUUCUUGCCAUAAAUUGGCUAAGCCAUUUACAGUU----GUGGCA ....(((((((((.....)))--------))))))...............((((((((......))))))))....((((.....))))...(((((........----))))). ( -32.80) >DroYak_CAF1 49803 103 - 1 UCCGUCUGGCAUUAAUUGAAU--------GCCAGAAUGCAAAACCAAAUCGAAUGCAUAAUUCGAUGCAUUUUCUUGCCAUAAAUUGGCUAAACCAUUUACAGUU----GUGGCA ....(((((((((.....)))--------))))))(((((........((((((.....))))))))))).....((((((((..(((.....))).......))----)))))) ( -31.10) >DroAna_CAF1 54420 110 - 1 UCCGUCUAGCAU----UGAAUGCCAGGGUGGCAGGUUUCUAGACAGUAUCGAAUAUUUAAUUCGUUGCAUUC-CUGGCCAUAAAUUGGCUAAGCAAAUUACCGUUCGUCAAAGCA ...((((((...----.(..((((.....))))..)..)))))).(((.(((((.....))))).)))....-.((((((.....))))))........................ ( -27.70) >consensus UCCGUCUGGCAUUAAUUGAAUGCAA___UGCCAGAAUGCAAAACCAAAUCGAAUGCAUAAUUCGAUGCAUUCUCUGGCCAUAAAUUGGCUAAACAAUUUACAGUU____GUGGCA ........(((((.....)))))......((((.................(((((((........)))))))..((((((.....))))))...................)))). (-15.69 = -17.13 + 1.45)


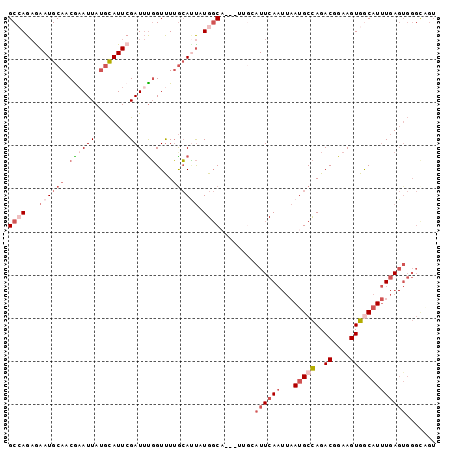
| Location | 8,268,162 – 8,268,258 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 80.34 |
| Mean single sequence MFE | -27.78 |
| Consensus MFE | -12.35 |
| Energy contribution | -15.02 |
| Covariance contribution | 2.67 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.44 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.578146 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8268162 96 + 22407834 GCCAGAGAAUGCAUCGAAUUAUGCAUUCGAUUUGGUUUUACACUAUGGCA---UUGCAUUCAAUUAAUGCCAGACGGAAGUGGCAUUUGAGUGGGCGGU ((((..((((((((......))))))))....((((.....)))))))).---...(((((((...(((((..((....))))))))))))))...... ( -31.40) >DroSec_CAF1 46203 96 + 1 GCCAGAGAAUGCAACAAAUUAUGCAUUCGAUUUGGUUUUGCAUUAUGGCA---UUGCAUUCAAUUAAUGCCAGACGGAAGUGACAUUUGAGUGGGCAGU (((((((((((((........)))))))..)))))).((((....(((((---(((........)))))))).((....)).............)))). ( -27.80) >DroSim_CAF1 46294 96 + 1 GCCAGAGAAUGCAACAAAUUAUGCAUUCGAUUUGGUUUUGCAUUAUGGCA---UUGCAUUCAAUUAAUGCCAGACGGAAGUGACAUUUGAGUGGGCAGU (((((((((((((........)))))))..)))))).((((....(((((---(((........)))))))).((....)).............)))). ( -27.80) >DroEre_CAF1 48497 91 + 1 GCAAGAAAAUGCAUCGCAUUAUGCAUUCGAUUUGGUUUUGCAUUCUGGC--------AUUCAAUUAAUGCCAGACGGAAGUGGCAUUUGAGUGGGUAGU (((......))).....(((((.(((((((........(((...(((((--------(((.....))))))))((....)).))).))))))).))))) ( -27.00) >DroYak_CAF1 49836 91 + 1 GCAAGAAAAUGCAUCGAAUUAUGCAUUCGAUUUGGUUUUGCAUUCUGGC--------AUUCAAUUAAUGCCAGACGGAAGUGGCAUUUGAGUGGGUUGU ((((...(((((((......)))))))(.(((..(...(((...(((((--------(((.....))))))))((....)).))).)..))).).)))) ( -26.30) >DroAna_CAF1 54457 85 + 1 GCCAG-GAAUGCAACGAAUUAAAUAUUCGAUACUGUCUAGAAACCUGCCACCCUGGCAUUCA----AUGCUAGACGGAAGUGGCUUAUAA--------- ((((.-........(((((.....)))))...((((((((.....((((.....))))....----...))))))))...))))......--------- ( -26.40) >consensus GCCAGAGAAUGCAACGAAUUAUGCAUUCGAUUUGGUUUUGCAUUAUGGCA___UUGCAUUCAAUUAAUGCCAGACGGAAGUGGCAUUUGAGUGGGCAGU ((((...((((((.((((((........))))))....)))))).))))........((((((...(((((..((....)))))))))))))....... (-12.35 = -15.02 + 2.67)


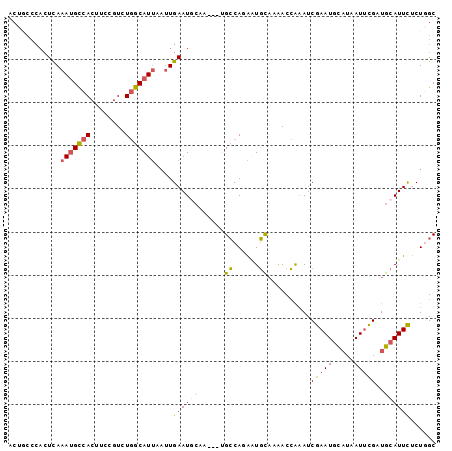
| Location | 8,268,162 – 8,268,258 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 80.34 |
| Mean single sequence MFE | -22.96 |
| Consensus MFE | -11.33 |
| Energy contribution | -11.58 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.886029 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8268162 96 - 22407834 ACCGCCCACUCAAAUGCCACUUCCGUCUGGCAUUAAUUGAAUGCAA---UGCCAUAGUGUAAAACCAAAUCGAAUGCAUAAUUCGAUGCAUUCUCUGGC ...(((......(((((((........)))))))....((((((.(---(((....))))........(((((((.....)))))))))))))...))) ( -25.50) >DroSec_CAF1 46203 96 - 1 ACUGCCCACUCAAAUGUCACUUCCGUCUGGCAUUAAUUGAAUGCAA---UGCCAUAAUGCAAAACCAAAUCGAAUGCAUAAUUUGUUGCAUUCUCUGGC ...(((......(((((((........)))))))....((((((((---(...((.(((((.............))))).))..)))))))))...))) ( -21.62) >DroSim_CAF1 46294 96 - 1 ACUGCCCACUCAAAUGUCACUUCCGUCUGGCAUUAAUUGAAUGCAA---UGCCAUAAUGCAAAACCAAAUCGAAUGCAUAAUUUGUUGCAUUCUCUGGC ...(((......(((((((........)))))))....((((((((---(...((.(((((.............))))).))..)))))))))...))) ( -21.62) >DroEre_CAF1 48497 91 - 1 ACUACCCACUCAAAUGCCACUUCCGUCUGGCAUUAAUUGAAU--------GCCAGAAUGCAAAACCAAAUCGAAUGCAUAAUGCGAUGCAUUUUCUUGC ..............(((........(((((((((.....)))--------))))))..)))....(((...((((((((......))))))))..))). ( -20.60) >DroYak_CAF1 49836 91 - 1 ACAACCCACUCAAAUGCCACUUCCGUCUGGCAUUAAUUGAAU--------GCCAGAAUGCAAAACCAAAUCGAAUGCAUAAUUCGAUGCAUUUUCUUGC ...........((((((........(((((((((.....)))--------))))))............(((((((.....)))))))))))))...... ( -24.00) >DroAna_CAF1 54457 85 - 1 ---------UUAUAAGCCACUUCCGUCUAGCAU----UGAAUGCCAGGGUGGCAGGUUUCUAGACAGUAUCGAAUAUUUAAUUCGUUGCAUUC-CUGGC ---------......((((.....((((((...----.(..((((.....))))..)..)))))).(((.(((((.....))))).)))....-.)))) ( -24.40) >consensus ACUGCCCACUCAAAUGCCACUUCCGUCUGGCAUUAAUUGAAUGCAA___UGCCAGAAUGCAAAACCAAAUCGAAUGCAUAAUUCGAUGCAUUCUCUGGC ............(((((((........)))))))....((((((......((......))..........(((((.....)))))..))))))...... (-11.33 = -11.58 + 0.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:38:36 2006