| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,262,271 – 8,262,384 |
| Length | 113 |
| Max. P | 0.832074 |

| Location | 8,262,271 – 8,262,384 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.38 |
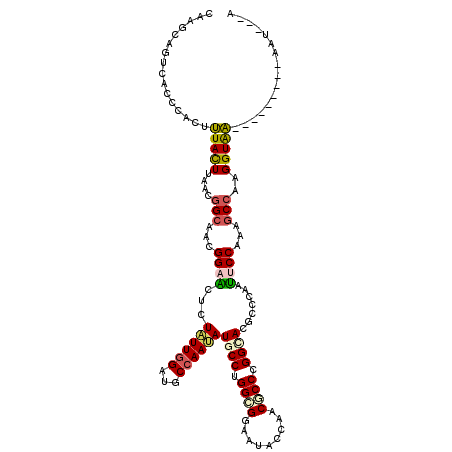
| Mean single sequence MFE | -27.79 |
| Consensus MFE | -19.00 |
| Energy contribution | -19.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.832074 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
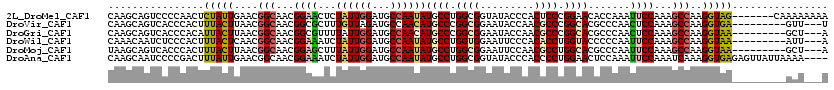
>2L_DroMel_CAF1 8262271 113 - 22407834 CAAGCAGUCCCCAACUCUAUUGAACGGCAACGGAACUCUAUUGGAUGCCAAUAUGCCUGGCGGUAUACCCACUCCCGGAACACCAAAUUCCAAAGCCAAGGUAG-------CAAAAAAAA ...((....................((((.(.((......)).).))))....((((((((((....)).......((((.......))))...))).))))))-------)........ ( -24.80) >DroVir_CAF1 50518 108 - 1 CAAGCAGUCACCCACUUUACUUAACGGCAACGGCGCUUUGUUAGAUGCCAACAUGCCCGGCGGAAUACCAACGCCCGGCACGCCCAACUCCAAAGCCAAGGUGA---------GUU---U .......(((((..((((.......(((...((((((.....)).))))....((((.((((.........)))).)))).))).......))))....)))))---------...---. ( -31.84) >DroGri_CAF1 53939 108 - 1 CAAGCAGUCACCCACAUUACUUAACGGCAACGGCGUUUUAUUGGAUGCCAACAUGCCCGGCGGAAUACCAACGCCCGGCACGCCCAACUCCAAAGCCAAGGUAA---------GCU---A ..(((.((.....)).((((((...(((...(((((((....)))))))....((((.((((.........)))).))))..............))).))))))---------)))---. ( -32.50) >DroWil_CAF1 46864 108 - 1 CAAACAAUCUCCCACUUUACUCAACGGCAACGGAAAUCUAUUGGAUGCCAAUAUGCCUGGUGGAAUUCCCACACCUGGUACCCCCAAUUCCAAAGCCAAGGUAA---------AUU---A ...............((((((....(((...((((...((((((...))))))((((.((((.........)))).)))).......))))...)))..)))))---------)..---. ( -24.70) >DroMoj_CAF1 50083 108 - 1 UAAGCAGUCACCCACUUUACUUAACGGCAACGGAGCUUUAUUGGAUGCCAAUAUGCCUGGCGGAAUUCCAACGCCUGGCACGCCCAAUUCCAAAGCCAAGGUAA---------GCU---A ..((((((.....)))((((((..((....))(.((((((((((..((.....((((.((((.........)))).)))).)))))))...)))))).))))))---------)))---. ( -30.10) >DroAna_CAF1 48840 116 - 1 CAAGCAAUCCCCGACUUUAUUGAACGGCAACGGAAAUCUAUUGGAUGCCAAUAUGCCUGGCGGUAUACCCACCCCUGGAACUCCAAAUUCCAAAUCAAAGGUGAGAGUUAUUAAAA---- ............((((((.......((((.(.((......)).).)))).....((((((.(((......)))))(((((.......)))))......)))).)))))).......---- ( -22.80) >consensus CAAGCAGUCACCCACUUUACUUAACGGCAACGGAACUCUAUUGGAUGCCAAUAUGCCUGGCGGAAUACCAACGCCCGGCACGCCCAAUUCCAAAGCCAAGGUAA_________AAU___A ................(((((....(((...((((...((((((...))))))((((.((((.........)))).)))).......))))...)))..)))))................ (-19.00 = -19.00 + 0.00)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:38:32 2006