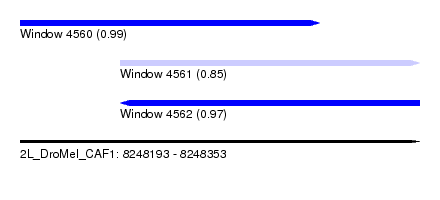
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,248,193 – 8,248,353 |
| Length | 160 |
| Max. P | 0.993046 |

| Location | 8,248,193 – 8,248,313 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.89 |
| Mean single sequence MFE | -42.85 |
| Consensus MFE | -27.41 |
| Energy contribution | -25.95 |
| Covariance contribution | -1.46 |
| Combinations/Pair | 1.66 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.64 |
| SVM decision value | 2.37 |
| SVM RNA-class probability | 0.993046 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
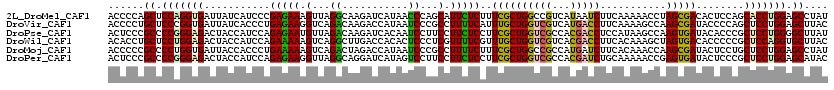
>2L_DroMel_CAF1 8248193 120 + 22407834 UGAAGGCAGGAUUAUGAUCCUGUCUGACUUUUUCAGGAGAAUAGGCUCCAGGUGCUGGAGUGUCGCUAGGUUUUUGAAGAUUAUGACGGCCAGCGAAAGAGAAUGCUGGGUUAUGAUCUU ...(((((((((....)))))))))......((((((((..((((((((((...)))))))....)))..))))))))(((((((((..((((((........))))))))))))))).. ( -45.70) >DroVir_CAF1 29141 120 + 1 UGAAUGCGGGAUUGUGAUCCUGACGGACUUUUUCAGGGGAGUAAGCUCCAGGAGCUGGGGUAUCGCUUGGCUUUUGAAGGUCAUGACGACCAGCAAAUGAAAAGGCGGGAUUAUGGUCUU .......(((((..((((((((.....((((((((..((((....))))....(((((....((((.((((((....)))))).).))))))))...))))))))))))))))..))))) ( -44.80) >DroPse_CAF1 33628 120 + 1 AAAAAGCUGGGUUAUGGUCUUGUCGGUUCUUUUCCGGAGAAUAAGCCCCAGGAGCGGGUGUAUCACUUGGCUUAUGGAGGUCGUGGCGACCAGCGAAGGAGAAGGAAGGAUUGUGAUCUU ........((((((..(((((.....(((((((((............(((.(((((((((...))))).)))).))).(((((...)))))......))))))))))))))..)))))). ( -39.00) >DroWil_CAF1 28948 120 + 1 AAAAGGCAGGAUUGUGAUCUUGUCUAACCUUUUCGGGAGAGUAAGCACCUGGAGCGGGGGUGUCACUAGCUUUGUGAAGGUCGUGACGACCAGCAAACGAAAACGAGGGAGUGUGGUCAA ...(((((((((....)))))))))..((.(((((((.(......).))))))).))....(((((.(.(((....))).).)))))(((((.((..(........)....))))))).. ( -35.50) >DroMoj_CAF1 31725 120 + 1 AAAAGGCAGGAUUGUGGUCUAGCCGAACUUUCUCUGGGGAAUAGGCUCCAGGAGCAGGAGUAUCGCUUGGUUUGUGAAGAUCAUGGCGGCCAGCGAAAGAAAAGGCGGGAUUAUGGUCUA ........((((..((((((.(((...(((((.((((.......(((((.......))))).(((((((((((....)))))).))))))))).)))))....))).))))))..)))). ( -46.70) >DroPer_CAF1 35233 120 + 1 UGUAGGAUGGAUUGUAAUCCUGCCGCACCUUCUCAGGAGAGUAUGCUCCAGGAGCGGGAGUAUCACUCGGUUUUUGCAGAUCGUGGCGACCAGCGAAGGAGAAGGAAGGACUAUGAUCCU ........((((..((.((((.((...(((((.(....(((((((((((.......))))))).))))((((.(..(.....)..).)))).).)))))....)).)))).))..)))). ( -45.40) >consensus AAAAGGCAGGAUUGUGAUCCUGUCGGACCUUUUCAGGAGAAUAAGCUCCAGGAGCGGGAGUAUCACUUGGUUUUUGAAGAUCAUGACGACCAGCGAAAGAAAAGGAGGGAUUAUGAUCUU ........(((((((((((((......((((((((((((......))))....((.((....(((..((((((....)))))))))...)).))...)))))))).))))))))))))). (-27.41 = -25.95 + -1.46)


| Location | 8,248,233 – 8,248,353 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.06 |
| Mean single sequence MFE | -43.52 |
| Consensus MFE | -26.38 |
| Energy contribution | -24.53 |
| Covariance contribution | -1.85 |
| Combinations/Pair | 1.58 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.851788 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8248233 120 + 22407834 AUAGGCUCCAGGUGCUGGAGUGUCGCUAGGUUUUUGAAGAUUAUGACGGCCAGCGAAAGAGAAUGCUGGGUUAUGAUCUUGCCUAACUUUCUCGGGAUGAUAAUCACCUGGAGCUGGGGU ...(((((((((((((.(((....(.(((((.....(((((((((((..((((((........)))))))))))))))))))))).)...))).))........)))))))))))..... ( -47.70) >DroVir_CAF1 29181 120 + 1 GUAAGCUCCAGGAGCUGGGGUAUCGCUUGGCUUUUGAAGGUCAUGACGACCAGCAAAUGAAAAGGCGGGAUUAUGGUCUUGUCUGACCUUCUCAGGGUGAUAAUCACCGGGAGCAGGGGU ....(((((((((((..((......))..))))))((((((((.((((((((.....((......))......)))))..)))))))))))....((((.....)))).)))))...... ( -47.30) >DroPse_CAF1 33668 120 + 1 AUAAGCCCCAGGAGCGGGUGUAUCACUUGGCUUAUGGAGGUCGUGGCGACCAGCGAAGGAGAAGGAAGGAUUGUGAUCUUGUCUAACAUUCUCUGGAUGGUAGUCUCCCGGGGCGGGAGU ....(((((..(((((((((...))))).))))..(((((.(...((.(((((.(((..(((....(((((....))))).)))....))).)))).).)).)))))).)))))...... ( -40.00) >DroWil_CAF1 28988 120 + 1 GUAAGCACCUGGAGCGGGGGUGUCACUAGCUUUGUGAAGGUCGUGACGACCAGCAAACGAAAACGAGGGAGUGUGGUCAAGCCUGACUUUUUCUGGAUGGUAGUCUCCAGGAGCAGGUGU ....((((((((((((((...(((((.(.(((....))).).)))))(((((.((..(........)....)))))))...)))).)))((((((((........))))))))))))))) ( -41.90) >DroMoj_CAF1 31765 120 + 1 AUAGGCUCCAGGAGCAGGAGUAUCGCUUGGUUUGUGAAGAUCAUGGCGGCCAGCGAAAGAAAAGGCGGGAUUAUGGUCUAGUCUGACUUUUUCAGGGUGGUAAUCACCAGGGGCGGGGGU ....((((((((((((((....((((.......))))((((((((((.(((..(....)....))).)..)))))))))..)))).)))))....((((.....)))).)))))...... ( -37.90) >DroPer_CAF1 35273 120 + 1 GUAUGCUCCAGGAGCGGGAGUAUCACUCGGUUUUUGCAGAUCGUGGCGACCAGCGAAGGAGAAGGAAGGACUAUGAUCCUGCCUAACCUUCUCUGGAUGGUAGUCUCCCGGGGCGGGAGU ....(((((.....((((((((((((.(((((......))))).)....((((.(((((...(((.((((......)))).)))..))))).)))).)))))..)))))).....))))) ( -46.30) >consensus AUAAGCUCCAGGAGCGGGAGUAUCACUUGGUUUUUGAAGAUCAUGACGACCAGCGAAAGAAAAGGAGGGAUUAUGAUCUUGCCUAACUUUCUCUGGAUGGUAAUCACCAGGAGCGGGGGU ....(((((.(((((.((....(((..((((((....)))))))))...)).))..............((((((.((((...............)))).))))))))).)))))...... (-26.38 = -24.53 + -1.85)


| Location | 8,248,233 – 8,248,353 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.06 |
| Mean single sequence MFE | -34.12 |
| Consensus MFE | -19.99 |
| Energy contribution | -18.93 |
| Covariance contribution | -1.05 |
| Combinations/Pair | 1.41 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.973384 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8248233 120 - 22407834 ACCCCAGCUCCAGGUGAUUAUCAUCCCGAGAAAGUUAGGCAAGAUCAUAACCCAGCAUUCUCUUUCGCUGGCCGUCAUAAUCUUCAAAAACCUAGCGACACUCCAGCACCUGGAGCCUAU ......((((((((((...........(((...((((((.(((((.((.(((((((..........)))))..)).)).)))))......))))))....)))...)))))))))).... ( -36.04) >DroVir_CAF1 29181 120 - 1 ACCCCUGCUCCCGGUGAUUAUCACCCUGAGAAGGUCAGACAAGACCAUAAUCCCGCCUUUUCAUUUGCUGGUCGUCAUGACCUUCAAAAGCCAAGCGAUACCCCAGCUCCUGGAGCUUAC ......(((((.((((.....))))(((.(((((((((((..((((((((..............))).)))))))).))))))))....((...)).......))).....))))).... ( -36.24) >DroPse_CAF1 33668 120 - 1 ACUCCCGCCCCGGGAGACUACCAUCCAGAGAAUGUUAGACAAGAUCACAAUCCUUCCUUCUCCUUCGCUGGUCGCCACGACCUCCAUAAGCCAAGUGAUACACCCGCUCCUGGGGCUUAU ......((((((((((......(((..(((((.(..((..............))..))))))(((.((((((((...)))))......))).))).))).......)))))))))).... ( -34.76) >DroWil_CAF1 28988 120 - 1 ACACCUGCUCCUGGAGACUACCAUCCAGAAAAAGUCAGGCUUGACCACACUCCCUCGUUUUCGUUUGCUGGUCGUCACGACCUUCACAAAGCUAGUGACACCCCCGCUCCAGGUGCUUAC ......((.(((((((((((.((..(.(((((.....((..((....))..))....))))))..)).)))))(((((...(((....)))...)))))........)))))).)).... ( -32.80) >DroMoj_CAF1 31765 120 - 1 ACCCCCGCCCCUGGUGAUUACCACCCUGAAAAAGUCAGACUAGACCAUAAUCCCGCCUUUUCUUUCGCUGGCCGCCAUGAUCUUCACAAACCAAGCGAUACUCCUGCUCCUGGAGCCUAU .....(((...(((((.........((((.....))))................(((............))))))))((.....))........)))...((((.......))))..... ( -19.40) >DroPer_CAF1 35273 120 - 1 ACUCCCGCCCCGGGAGACUACCAUCCAGAGAAGGUUAGGCAGGAUCAUAGUCCUUCCUUCUCCUUCGCUGGUCGCCACGAUCUGCAAAAACCGAGUGAUACUCCCGCUCCUGGAGCAUAC .((((((...)))))).......(((((.(((((..(((.(((((....))))).)))...)))))((.(((((...))))).)).......(((((.......))))))))))...... ( -45.50) >consensus ACCCCCGCCCCGGGAGACUACCAUCCAGAGAAAGUCAGACAAGACCAUAAUCCCGCCUUCUCCUUCGCUGGUCGCCACGACCUUCAAAAACCAAGCGAUACUCCCGCUCCUGGAGCUUAC ......((.(((((((...........(((((.(...((...........))...).)))))..((((((((((...)))))...........)))))........))))))).)).... (-19.99 = -18.93 + -1.05)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:38:21 2006