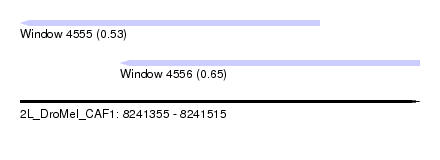
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,241,355 – 8,241,515 |
| Length | 160 |
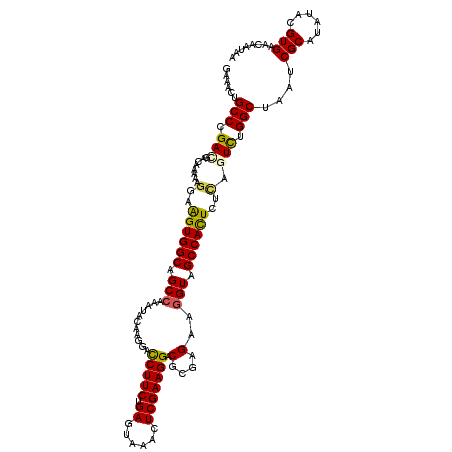
| Max. P | 0.650533 |

| Location | 8,241,355 – 8,241,475 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.50 |
| Mean single sequence MFE | -33.22 |
| Consensus MFE | -22.94 |
| Energy contribution | -24.34 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.69 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.528990 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8241355 120 - 22407834 AAGGACCUUCUGAGUAAACUCGAAGGACGCGAGAAGGUAGCCACUCUCAGUUUGGCUAAUCGCAUAUACGUGAACAAAAAGUUCCAGCUAGUACCGAGUUACAAUCAGAUGGUCAAGGAU ...((((.(((((...((((((....((((((.....((((((.........)))))).))))......(.((((.....))))).....))..))))))....))))).))))...... ( -37.30) >DroSec_CAF1 19471 120 - 1 AAGGACCUUCUGAGCAAAAUCGAAGGACGCGAGAAGGUAGCCACUCUCAGUCUGGCUAAUCGCAUAUACGUGAACGAUAGGUUCACGCUAAUACCAAAGUACAAUCAGAUGGUCAAGGAU ...((((.(((((((....((....)).((((.....((((((.........)))))).))))......((((((.....))))))))...(((....)))...))))).))))...... ( -36.50) >DroSim_CAF1 19613 120 - 1 AAGGACCUUCUGAGUAAAAUCGAAGGACGCGAGAAGGUAGCCACUCUCAGUCUGGCUAAUCGCAUAUACGUGAACGAUAAGUUCAAGCUAGUACCGAAGUACAAUCAGAUGGUCAAGGAU ...((((.((((((((...((....)).((((.....((((((.........)))))).))))...))).(((((.....))))).....((((....))))..))))).))))...... ( -36.40) >DroEre_CAF1 20611 120 - 1 AAGGAUCUUCUGACUGAACUCGAAGGUCGUGAGAAAGUAGCCAUUCUUACUCUGGCGAACCGCAUAUACGUGAACAAUAAGAUAAAGCUAUUACCGCAGUACAAUCAGGUGGUCAAGGAU ...((((.(((((.((.(((((....((....)).((((((...(((((.....(((...)))......(....)..)))))....))))))..)).))).)).))))).))))...... ( -28.50) >DroYak_CAF1 22972 120 - 1 AAGGAUCUUCUGACGGAGCUCGAAGGUCGCGAGAAGGUAGCCAUUCUAAGUCUGGCAAACCGCAUAUACGUGAACAAUAAGUUCGAACUAGUACCAGGGUACAAUCAGAUAGUGAAGGAU ....((..(((((..(((((......(((((....(((.((((.........))))..))).......)))))......)))))......((((....))))..)))))..))....... ( -27.40) >consensus AAGGACCUUCUGAGUAAACUCGAAGGACGCGAGAAGGUAGCCACUCUCAGUCUGGCUAAUCGCAUAUACGUGAACAAUAAGUUCAAGCUAGUACCGAAGUACAAUCAGAUGGUCAAGGAU ...((((.(((((...........((((((((.....((((((.........)))))).))))......(....).....))))......((((....))))..))))).))))...... (-22.94 = -24.34 + 1.40)

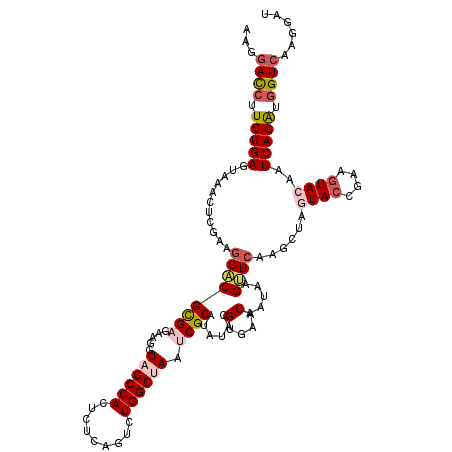
| Location | 8,241,395 – 8,241,515 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.58 |
| Mean single sequence MFE | -32.90 |
| Consensus MFE | -25.96 |
| Energy contribution | -25.76 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.650533 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8241395 120 - 22407834 GAAACUGCCCGAUGACAAAAAGGAAGUGGCAGCCAAAUACAAGGACCUUCUGAGUAAACUCGAAGGACGCGAGAAGGUAGCCACUCUCAGUUUGGCUAAUCGCAUAUACGUGAACAAAAA .........((((..((((..(..((((((.(((...........(((((.(((....)))))))).(....)..))).))))))..)..))))....)))).................. ( -34.80) >DroSec_CAF1 19511 120 - 1 GAAACUGCCCGACGACAAAAAGGAAGUGGCAGCCAAAUACAAGGACCUUCUGAGCAAAAUCGAAGGACGCGAGAAGGUAGCCACUCUCAGUCUGGCUAAUCGCAUAUACGUGAACGAUAG ......(((.(((........(..((((((..((........))(((((((..((....((....)).)).))))))).))))))..).))).)))...((((......))))....... ( -35.10) >DroSim_CAF1 19653 120 - 1 GAAACUGCCCGACGACAAAAAGGAAGUGGCAGCCAAAUACAAGGACCUUCUGAGUAAAAUCGAAGGACGCGAGAAGGUAGCCACUCUCAGUCUGGCUAAUCGCAUAUACGUGAACGAUAA ......(((.(((........(..((((((.(((...........(((((.((......))))))).(....)..))).))))))..).))).)))...((((......))))....... ( -34.70) >DroEre_CAF1 20651 120 - 1 GGGACUGCCCGAUAACAGGGAGGAGGUGGCAGCCAAAACCAAGGAUCUUCUGACUGAACUCGAAGGUCGUGAGAAAGUAGCCAUUCUUACUCUGGCGAACCGCAUAUACGUGAACAAUAA (((....))).....((.(......((((..((((........(((((((.((......)))))))))(((((((........)))))))..))))...)))).....).))........ ( -32.70) >DroYak_CAF1 23012 120 - 1 GAAACUGCCCGAGGACAAAAAGCAAGUGGCAGCCAAGUACAAGGAUCUUCUGACGGAGCUCGAAGGUCGCGAGAAGGUAGCCAUUCUAAGUCUGGCAAACCGCAUAUACGUGAACAAUAA .....((((...((((....((..((((((.(((.........((.((((....)))).)).....((....)).))).))))))))..))))))))...(((......)))........ ( -27.20) >consensus GAAACUGCCCGACGACAAAAAGGAAGUGGCAGCCAAAUACAAGGACCUUCUGAGUAAACUCGAAGGACGCGAGAAGGUAGCCACUCUCAGUCUGGCUAAUCGCAUAUACGUGAACAAUAA ......(((.(((........(..((((((.(((...........(((((.((......))))))).(....)..))).))))))..).))).)))....(((......)))........ (-25.96 = -25.76 + -0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:38:15 2006