| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,193,141 – 8,193,256 |
| Length | 115 |
| Max. P | 0.879719 |

| Location | 8,193,141 – 8,193,256 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
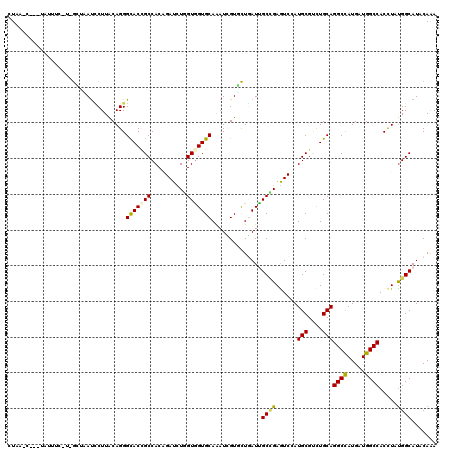
| Mean pairwise identity | 78.97 |
| Mean single sequence MFE | -38.10 |
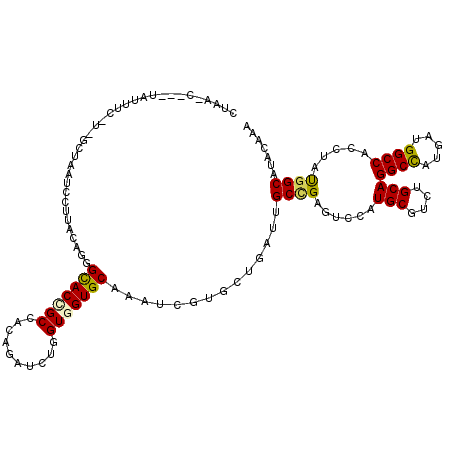
| Consensus MFE | -23.79 |
| Energy contribution | -23.68 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.879719 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
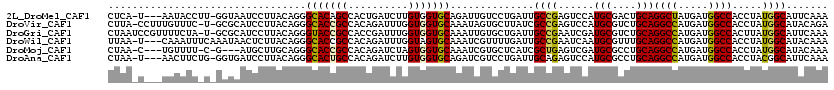
>2L_DroMel_CAF1 8193141 115 - 22407834 CUCA-U---AAUACCUU-GGUAAUCCUUACAGGGCACAGCCACUGAUCUUGUGGUGCAGAUUGUCCUGAUUGCCGAGUCCAUGCGACUGCAGGCUAUGAUGGCCACCUAUGGCAUUCAAA .(((-(---(....(((-(((((((.....((((((..(((((.......)))))......)))))))))))))))).((.(((....))))).)))))..((((....))))....... ( -36.50) >DroVir_CAF1 39196 117 - 1 CUUA-CCUUUGUUUC-U-GCGCAUCCUUACAGGGCACCGCCACAGAUUUGGUGGUGCAAAUAGUGCUUAUCGCCGAGUCCAUGCGUCUGCAGGCCAUGAUGGCCACCUAUGGCAUACAGA ....-..(((((...-.-((((..((.....))(((((((((......))))))))).....)))).....((((......(((....)))((((.....)))).....))))..))))) ( -40.20) >DroGri_CAF1 41569 118 - 1 CUAAUCCGUUUUCUA-U-GCGCAUCCUUACAGGGUACCGCCACCGAUUUGGUGGUGCAAAUUGUGCUGAUUGCCGAAUCGAUGCGUCUGCAGGCCAUGAUGGCCACUUAUGGCAUUCAAA ...............-.-(((((.((.....))(((((((((......)))))))))....)))))(((.(((((......(((....)))((((.....)))).....))))).))).. ( -41.50) >DroWil_CAF1 27530 116 - 1 UUAA-U---CAAAUUUCAAAUAACUCUUACAGGGCACCGCCACAGAUUUGGUAGUGCAAAUCGUUUUGAUUGCCGAAUCAAUGCGUUUGCAGGCCAUGAUGGCCACCUAUGGCAUACAAA ....-.---.....................((((((.(((....(((((((((((.((((....)))))))))))))))...)))..))).((((.....)))).)))............ ( -32.40) >DroMoj_CAF1 39549 111 - 1 CUAA-C---UGUUUU-C-G---AUGCUUGCAGGGCACCGCCACAGAUCUAGUGGUGCAAAUCGUGCUCAUCGCUGAGUCGAUGCGCCUGCAGGCCAUGAUGGCCACCUAUGGCAUACAAA ....-.---......-.-.---((((((((((((((((((..........)))))))..((((.(((((....)))))))))...))))))((((.....))))......)))))..... ( -42.00) >DroAna_CAF1 19604 115 - 1 CUAA-U---AACUUCUG-GGUGAUCCUUACAGGGCACUGCCACAGAUCUUGUGGUGCAGAUCGUCCUGAUUGCAGAGUCCAUGCGCCUGCAGGCCAUGAUGGCCACCUACGGCAUUCAAA ....-.---......((-((((..((...((((((.(((((((((...)))))..))))...))))))..(((((.((......)))))))((((.....))))......)))))))).. ( -36.00) >consensus CUAA_C___UAUUUC_U_GCUAAUCCUUACAGGGCACCGCCACAGAUCUGGUGGUGCAAAUCGUGCUGAUUGCCGAGUCCAUGCGUCUGCAGGCCAUGAUGGCCACCUAUGGCAUACAAA .................................(((((((..........)))))))..............((((......(((....)))((((.....)))).....))))....... (-23.79 = -23.68 + -0.11)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:38:05 2006