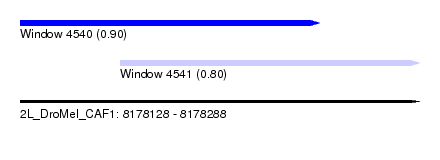
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,178,128 – 8,178,288 |
| Length | 160 |
| Max. P | 0.900428 |

| Location | 8,178,128 – 8,178,248 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.94 |
| Mean single sequence MFE | -44.05 |
| Consensus MFE | -33.11 |
| Energy contribution | -32.00 |
| Covariance contribution | -1.11 |
| Combinations/Pair | 1.35 |
| Mean z-score | -3.26 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.900428 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
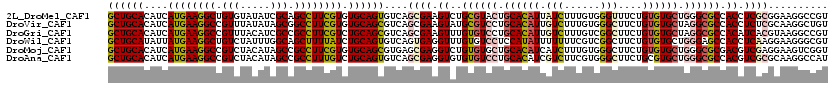
>2L_DroMel_CAF1 8178128 120 + 22407834 UCUUCAAGAACUUCAAGGAUGUCUUCUGGCGCUUCCUGGAGCUGCACAUCAUGAAGGCUGUGUAUAUCGCAGCCUUCGUGUGCAGUGUCAGCGAAGUCUGCGUACUGCACAUUAUCUUUG .............((((((((((....)))((((((((..(((((((((...((((((((((.....)))))))))))))))))))..))).))))).(((.....)))....))))))) ( -49.60) >DroVir_CAF1 21764 120 + 1 UAUUCAAGCGUUUCAAGACCGUCUUCUGGCGCUUCCUGGAGCUGCACAUCAUGAAGGCCGUUUAUAUAGCGGCCUUCGUGUGCAGCGUCAGCGAAGUAUGCGUCCUGCACAUUGUCUUUG ..............(((((.(((....)))((((((((..(((((((((...((((((((((.....)))))))))))))))))))..))).))))).(((.....)))....))))).. ( -51.60) >DroGri_CAF1 24166 120 + 1 UCUUCAAGAGAUUCAAGACCGUCUUUUGGCGCUUCCUCGAGCUGCACAUCAUGAAGGCCGUUUACAUCGCCGCCUUCGUCUGCAGCGUCAGCGAAGUUUGUGUCCUGCACAUUGUCUUUG ...(((((((((........))))))))).(((((((.((((((((....((((((((.((.......)).)))))))).)))))).)))).))))).((((.....))))......... ( -38.30) >DroWil_CAF1 5788 120 + 1 UAUUCAAGAAUUUCAAAGAUGUCUUCUGGCGUUUUCUUGAGCUGCAUAUUAUGAAGGCUGUCUAUUUGGCAGCUUUUAUCUGCAGUGUCAGUGAGGUUUGUGUCCUCCAUAUUUUUUUCG ...(((((((.......((((((....)))))))))))))((((((....((((((((((((.....)))))))))))).))))))....(.((((.......)))))............ ( -38.01) >DroMoj_CAF1 21773 120 + 1 UAUUCAAGCGCUUCAAGACCGUCUUCUGGCGCUUCCUCGAGCUGCACAUCAUGAAGGCCGUCUACAUAGCCGCCUUCGUGUGCAGCGUGAGCGAGGUCUGUGUGCUGCACAUCAUCUUUG ......(((((....((((((((....)))...((((((.(((((((((...((((((.((.......)).))))))))))))))).)))).)))))))..))))).............. ( -48.50) >DroAna_CAF1 5024 120 + 1 UAUUCAAGAACUUCAAGGAUGUCUUUUGGCGCUUCCUGGAGCUGCACAUCAUGAAGGCCGUCUACAUAGCCGCCUUUGUCUGCAGUGUCAGCGAGGUGUGUGUCCUGCACAUCGUCUUCG ...............((((((.......((((((((((..((((((....(..(((((.((.......)).)))))..).))))))..))).)))))))(((.....)))..)))))).. ( -38.30) >consensus UAUUCAAGAACUUCAAGAACGUCUUCUGGCGCUUCCUGGAGCUGCACAUCAUGAAGGCCGUCUACAUAGCCGCCUUCGUCUGCAGCGUCAGCGAAGUCUGUGUCCUGCACAUUGUCUUUG ..............((((..(((....)))(((((((((.((((((....((((((((.(((.....))).)))))))).)))))).)))).))))).((((.....))))...)))).. (-33.11 = -32.00 + -1.11)


| Location | 8,178,168 – 8,178,288 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.72 |
| Mean single sequence MFE | -49.65 |
| Consensus MFE | -36.70 |
| Energy contribution | -37.62 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.802820 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8178168 120 + 22407834 GCUGCACAUCAUGAAGGCUGUGUAUAUCGCAGCCUUCGUGUGCAGUGUCAGCGAAGUCUGCGUACUGCACAUUAUCUUUGUGGGUUUCUGUGUGCUGGGCGCCACCUCGCGGAAGGCCGU (((((((((...((((((((((.....)))))))))))))))))))....((((.((..((((...((((((.((((....))))....))))))...)))).)).))))((....)).. ( -55.20) >DroVir_CAF1 21804 120 + 1 GCUGCACAUCAUGAAGGCCGUUUAUAUAGCGGCCUUCGUGUGCAGCGUCAGCGAAGUAUGCGUCCUGCACAUUGUCUUUGUGGGCUUCUGUGUGCUAGGCGCCACCUCUCGCAAGGCUGU (((((((((...((((((((((.....)))))))))))))))))))(((.((((((...(((((..((((((.((((....))))....))))))..)))))...)).))))..)))... ( -60.80) >DroGri_CAF1 24206 120 + 1 GCUGCACAUCAUGAAGGCCGUUUACAUCGCCGCCUUCGUCUGCAGCGUCAGCGAAGUUUGUGUCCUGCACAUUGUCUUUGUCGGCUUCUGUGUGCUAGGCGCCACAUCACGUAAGGCCGU ((((((....((((((((.((.......)).)))))))).))))))....((((((..((((.....))))....))))))((((...((((.((.....))))))...(....))))). ( -42.20) >DroWil_CAF1 5828 120 + 1 GCUGCAUAUUAUGAAGGCUGUCUAUUUGGCAGCUUUUAUCUGCAGUGUCAGUGAGGUUUGUGUCCUCCAUAUUUUUUUCGUCGGCUUCUGUGUGCUGGGAGCCACCUCAAGGAAGGGCGU ((((((....((((((((((((.....)))))))))))).)))))).((..((((((.((((.....))))...........(((((((.......)))))))))))))..))....... ( -43.90) >DroMoj_CAF1 21813 120 + 1 GCUGCACAUCAUGAAGGCCGUCUACAUAGCCGCCUUCGUGUGCAGCGUGAGCGAGGUCUGUGUGCUGCACAUCAUCUUUGUGGGCUUCUGUGUGCUGGGCGCGACGUCGAGGAAGUCGGU (((((((((...((((((.((.......)).))))))))))))))).....(((.(((.((((.(.((((((..(((....))).....)))))).).))))))).)))........... ( -47.50) >DroAna_CAF1 5064 120 + 1 GCUGCACAUCAUGAAGGCCGUCUACAUAGCCGCCUUUGUCUGCAGUGUCAGCGAGGUGUGUGUCCUGCACAUCGUCUUCGUGGGCUUCUGCGUGCUGGGCGCCACGUCGCGCAAGGCCAU ((((((....(..(((((.((.......)).)))))..).))))))(((.((((.(((.((((((.((((..((....))...((....)))))).))))))))).))))....)))... ( -48.30) >consensus GCUGCACAUCAUGAAGGCCGUCUACAUAGCCGCCUUCGUCUGCAGCGUCAGCGAAGUCUGUGUCCUGCACAUUGUCUUUGUGGGCUUCUGUGUGCUGGGCGCCACCUCGCGGAAGGCCGU ((((((....((((((((.(((.....))).)))))))).))))))....((((.((..((((((.((((((.(((......)))....)))))).)))))).)).)))).......... (-36.70 = -37.62 + 0.92)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:38:00 2006