
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,173,578 – 8,173,678 |
| Length | 100 |
| Max. P | 0.838909 |

| Location | 8,173,578 – 8,173,678 |
|---|---|
| Length | 100 |
| Sequences | 6 |
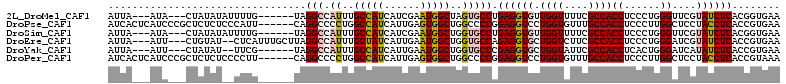
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 77.31 |
| Mean single sequence MFE | -28.78 |
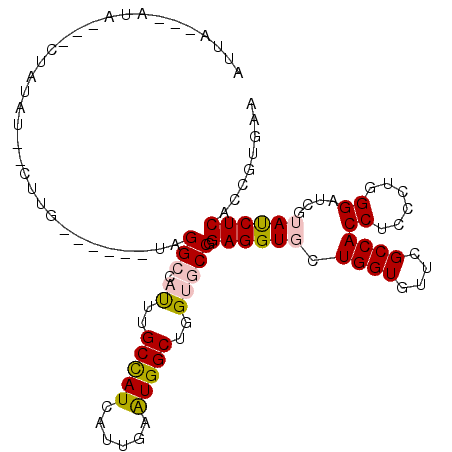
| Consensus MFE | -20.74 |
| Energy contribution | -20.93 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.838909 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8173578 100 + 22407834 AUUA---AUA---CUAUAUAUUUUG------UAGGCCAUUUGCCAUCAUCGAAUGGCUAGUGCCUGAGGUGUUGGUUUUCGCCACCUCCCUGGGUUCGUAUCUCACGGUGAA ....---...---(((((.....))------)))((((((((.......))))))))....((((((((((.((((....))))((.....)).....))))))).)))... ( -27.20) >DroPse_CAF1 438 106 + 1 AUCACUCAUCCCGCUCUCUCCCAUU------CAGGCCCCUGGCCAUCAUUGAGUGGCUGGCCCCGGAGGUCCUGGUGUUUGCCACCUCCCUUGGCUCCUACCUCACCGUGAA .((((.....(((...((..(((((------((((((...)))).....)))))))..))...))).(((...((((...((((.......))))...))))..))))))). ( -34.20) >DroSim_CAF1 448 100 + 1 AUUA---AUA---CUAUAUAUUUUG------UAGGCCAUUUGCCAUCAUCGAAUGGCUGGUGCCUGAGGUGUUGGUUUUCGCCACCUCCCUGGGUUCGUAUCUCACGGUGAA ....---...---(((((.....))------)))((((...(((((......)))))))))((((((((((.((((....))))((.....)).....))))))).)))... ( -27.40) >DroEre_CAF1 438 104 + 1 AUUA---AUU---CUGUAU--CUCAUUUGCUUAGGCCAUUUGCUAUCAUUGAAUGGCUGGUGCCAGAGGUGCUGGUCUUCGCCACCUCCCUGGGAUCGUAUCUCACCGUGAA ....---...---......--.((((.......((((((((.........))))))))(((.((((.((.(.((((....)))).).)))))).)))..........)))). ( -28.10) >DroYak_CAF1 464 98 + 1 AUUA---AUU---CUAUAU--UUCG------UAGGCCAUUUGCCAUCAUUGAAUGGCUGGUGCCCGAGGUGCUGGUAUUCGCCACCUCACUGGGAUCAUAUCUCACCGUGAA ....---...---...(((--((((------..(((.((..(((((......)))))..)))))))))))).((((....))))..((((((((((...))))))..)))). ( -26.20) >DroPer_CAF1 442 106 + 1 AUCACUCAUCCCGCUCUCUCCCCUU------CAGGCCCCUGGCCAUCAUUGAGUGGCUGGCCCCGGAGGUCCUGGUGUUUGCCACCUCCCUUGGCUCCUACCUCACCGUAAA .....................((((------(.((((...((((((......))))))))))..)))))....((((...((((.......))))...)))).......... ( -29.60) >consensus AUUA___AUA___CUAUAU__CUUG______UAGGCCAUUUGCCAUCAUUGAAUGGCUGGUGCCGGAGGUGCUGGUGUUCGCCACCUCCCUGGGAUCGUAUCUCACCGUGAA .................................(((.((..(((((......)))))..))))).((((((.((((....))))((......))....))))))........ (-20.74 = -20.93 + 0.20)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:37:57 2006