
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,167,843 – 8,168,001 |
| Length | 158 |
| Max. P | 0.744922 |

| Location | 8,167,843 – 8,167,944 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 78.76 |
| Mean single sequence MFE | -23.02 |
| Consensus MFE | -8.78 |
| Energy contribution | -9.66 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.38 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.744922 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
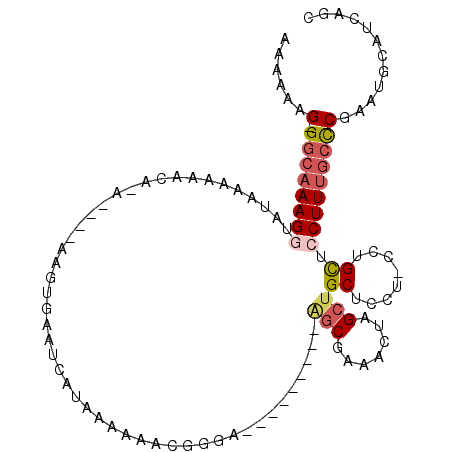
>2L_DroMel_CAF1 8167843 101 + 22407834 AAAAAAGGGGCAAAGGUAUAAAAAAAAAAUA-AAAGUGAAUCAUAAAAAACGGGA---------AGCGAAACUAGCUGCUCCUGCCUGCUCCUUUGCCCGAAUGCAUCAGC .......(((((((((...............-...(((...)))......(((((---------(((.......)))..)))))......)))))))))............ ( -23.50) >DroSec_CAF1 7709 94 + 1 AAAAAAG-GGAAAAGGUAUAAAAAACA------AAGUGAAUCAUAAAAAACGGGA---------GGCGA-ACUAGCAGCUCCUACCUGCUCCUUCGGCCGAAUGCAUCAGC .....((-(((..((((..........------..((............))((((---------(((..-....))..)))))))))..)))))..((.....))...... ( -17.00) >DroSim_CAF1 8233 95 + 1 AAAAAAG-GGCAAAGGUAUAAAAAACA------AAGUGAAUCAUAAAAAACGGGA---------GGCGAAACUAGCUGCUCCUGCCUGCUCCUUUGCUCGAAUGCAUCAGC ......(-((((((((...........------..(((...)))......(((((---------(((.......))..))))))......)))))))))............ ( -21.70) >DroEre_CAF1 8196 94 + 1 A---AAG-GGCAAAGACAUAAAAAGUCCAUGUGAAGUAAACCAUAAAAAAGGGGA---------AGCGAAACUAGCUGCUCC----UGUUCCUUUGCCCGAAUGCAUCAGC .---..(-(((((((..............((((........))))....(((((.---------(((.......))).))))----)....))))))))............ ( -25.90) >DroYak_CAF1 7907 104 + 1 AA--AAG-GGCAAAGAUAUAAAAGGUCCACGUGAAGUAAACCAUAAAAAACGGGAAAACGGGGUAGCGAAACUAGAUGCUAC----UGUUCCUUUGCCCGAAUGCAUCAGC ..--..(-(((((((........(.(((.(((....((.....))....))))))...)..(((((((........))))))----)....))))))))............ ( -27.00) >consensus AAAAAAG_GGCAAAGGUAUAAAAAACA_A____AAGUGAAUCAUAAAAAACGGGA_________AGCGAAACUAGCUGCUCCU_CCUGCUCCUUUGCCCGAAUGCAUCAGC ......(.((((((((................................................(((.......)))((........)).)))))))))............ ( -8.78 = -9.66 + 0.88)

| Location | 8,167,904 – 8,168,001 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.35 |
| Mean single sequence MFE | -32.23 |
| Consensus MFE | -12.19 |
| Energy contribution | -13.58 |
| Covariance contribution | 1.39 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.38 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8167904 97 - 22407834 AGG-----------GCAAGAAGCAAACUGG-CAUA-----------GCUCCUGCCUUGUUGUCCAUUAACAAACAAUUUGGCUGAUGCAUUCGGGCAAAGGAGCAGGCAGGAGCAGCUAG ...-----------((.....))...((((-(...-----------(((((((((((((((.....))))))....((((.((((.....)))).))))......))))))))).))))) ( -37.30) >DroSec_CAF1 7763 97 - 1 AGG-----------GCAAGAAGCAAGUUGG-CAUA-----------GCUCCUGCCUUGUUGUCCAUUAACAAACAAUUUGGCUGAUGCAUUCGGCCGAAGGAGCAGGUAGGAGCUGCUAG ..(-----------((.....((......)-)..(-----------(((((((((((((((.....))))))....(((((((((.....)))))))))......))))))))))))).. ( -39.20) >DroSim_CAF1 8288 97 - 1 AGG-----------GCAAGAAGCAAAAUGG-CAUA-----------GCUCCUGCCUUGUUGUCCAUUAACAAACAAUUUGGCUGAUGCAUUCGAGCAAAGGAGCAGGCAGGAGCAGCUAG ..(-----------((.....((......)-)...-----------((((((((((.((..(((.....(((.....)))(((((.....)).)))...))))))))))))))).))).. ( -33.70) >DroEre_CAF1 8254 92 - 1 -GG-----------ACAGGGAGCAAAACGG-CGUA-----------GAUCCUGCCUUGUUGUCCAUUAACAAACAAUUUGGCUGAUGCAUUCGGGCAAAGGAACA----GGAGCAGCUAG -..-----------..............((-(...-----------..(((((((((((((.....))))))....((((.((((.....)))).))))))..))----)))...))).. ( -22.40) >DroYak_CAF1 7975 95 - 1 -CG-----------GCGAGGAGCCAAACGG-CAUU------GC--UGCUCCUGCCUUGUUGUCCAUUAACAAACAAUUUGGCUGAUGCAUUCGGGCAAAGGAACA----GUAGCAUCUAG -..-----------....((((((....))-)..(------((--((((....((((((((.....))))))....((((.((((.....)))).))))))...)----))))))))).. ( -27.80) >DroAna_CAF1 9285 116 - 1 GGGGGAGUGUGAGAGGAAAGAGCGAAAGGAAGACAGCUCCUGCUAUGCUCCUCCUAUGCUGUCCAUUAACAAACAAUUUUGCUGAUGCAUUCGGCCCAAAGAACC----GAAACAGUUAG (((((((((((..((((....((............))))))..)))))))))))...(((((.(((((.((((....)))).)))))..(((((.........))----))))))))... ( -33.00) >consensus AGG___________GCAAGAAGCAAAAUGG_CAUA___________GCUCCUGCCUUGUUGUCCAUUAACAAACAAUUUGGCUGAUGCAUUCGGGCAAAGGAACA____GGAGCAGCUAG ..............................................(((((....((((((.....))))))....((((.((((.....)))).))))..........)))))...... (-12.19 = -13.58 + 1.39)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:37:53 2006