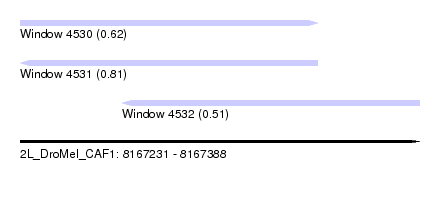
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,167,231 – 8,167,388 |
| Length | 157 |
| Max. P | 0.805489 |

| Location | 8,167,231 – 8,167,348 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.10 |
| Mean single sequence MFE | -31.61 |
| Consensus MFE | -27.04 |
| Energy contribution | -27.08 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.620141 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8167231 117 + 22407834 UGUACGUCAUUUGACAAGCCCCACCGUCAUUUGAAUAAAAUGCGCCCCGUUUCAGGGCCAAAACCUGUUGACUCAACUAUGUC---CUAUCCCUCGGGUCGAGUUGAGUCAACUAUUUGG .(((..(((..((((..........))))..)))......)))((((.......))))((((....((((((((((((.((.(---((.......))).))))))))))))))..)))). ( -35.90) >DroSec_CAF1 7107 116 + 1 UGUACGUCAUUUGACAAGCCCCACCUUCAUUUGAAUAAAAUGCGCCCCGUUUCAGGGCCAAAACCUGUUGACUCAACUAUG----UGUAUCCCUCGGGUCGAGUUGAGUCAACUAUUUGG .....(((....)))...............(..((((...((.((((.......))))))......((((((((((((.((----.....((....)).))))))))))))))))))..) ( -31.50) >DroSim_CAF1 7624 120 + 1 UGUACGUCAUUUGACAAGCCCCACCUUCAUUUGAAUAAAUUGUGCCCCGUUUCAGGGCCAAAACCUGUUGACUCAACUAUGUAUGUAUAUCCCUCGGGUCGAGUUGAGUCAACUAUUUGG .(((((((....)))..........(((....)))......))))(((......)))(((((....((((((((((((............((....))...))))))))))))..))))) ( -31.96) >DroEre_CAF1 7620 112 + 1 UGUACGUCCUUUGACAAGCCCCACCUUCGUUUAAAUGAAAUGCGCUCCGUUUCAGGGCCAAAACCUGUUGACUCAACU--------GUAUCCCUCGGGUCGAGCUGAGCCAACUAUUUGG .....(((((..(((.(((..((..(((((....))))).)).)))..)))..)))))((((....((((.((((.((--------..((((...))))..)).)))).))))..)))). ( -25.30) >DroYak_CAF1 7287 115 + 1 UGUACGUCAUUUGACAAGCCCCACCUUCAUUUGAAUGAAAUGCGCCCCGUUUCAGGGCCAAAACCUGUUGCCUCAGCUA-G----UAUAUCCCUCGGGUCGGGCUGAGUCAACUAUUUGG .....(((....)))..((((....(((....)))(((((((.....)))))))))))((((....((((.(((((((.-.----.....((....))...))))))).))))..)))). ( -33.40) >consensus UGUACGUCAUUUGACAAGCCCCACCUUCAUUUGAAUAAAAUGCGCCCCGUUUCAGGGCCAAAACCUGUUGACUCAACUAUG____UAUAUCCCUCGGGUCGAGUUGAGUCAACUAUUUGG .....(((....))).....(((.................((.((((.......))))))......((((((((((((............((....))...))))))))))))....))) (-27.04 = -27.08 + 0.04)



| Location | 8,167,231 – 8,167,348 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.10 |
| Mean single sequence MFE | -35.24 |
| Consensus MFE | -30.54 |
| Energy contribution | -30.62 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.805489 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8167231 117 - 22407834 CCAAAUAGUUGACUCAACUCGACCCGAGGGAUAG---GACAUAGUUGAGUCAACAGGUUUUGGCCCUGAAACGGGGCGCAUUUUAUUCAAAUGACGGUGGGGCUUGUCAAAUGACGUACA .......((((((((((((....((........)---)....))))))))))))(((((..(((((((...))))))((((((.....))))).)..)..)))))(((....)))..... ( -37.20) >DroSec_CAF1 7107 116 - 1 CCAAAUAGUUGACUCAACUCGACCCGAGGGAUACA----CAUAGUUGAGUCAACAGGUUUUGGCCCUGAAACGGGGCGCAUUUUAUUCAAAUGAAGGUGGGGCUUGUCAAAUGACGUACA .......((((((((((((...((....)).....----...))))))))))))(((((..(((((((...)))))).(((((.....)))))....)..)))))(((....)))..... ( -34.80) >DroSim_CAF1 7624 120 - 1 CCAAAUAGUUGACUCAACUCGACCCGAGGGAUAUACAUACAUAGUUGAGUCAACAGGUUUUGGCCCUGAAACGGGGCACAAUUUAUUCAAAUGAAGGUGGGGCUUGUCAAAUGACGUACA (((....(((((((((((....((....)).(((......)))))))))))))).....(((((((((...)))))).)))................))).(..((((....))))..). ( -34.60) >DroEre_CAF1 7620 112 - 1 CCAAAUAGUUGGCUCAGCUCGACCCGAGGGAUAC--------AGUUGAGUCAACAGGUUUUGGCCCUGAAACGGAGCGCAUUUCAUUUAAACGAAGGUGGGGCUUGUCAAAGGACGUACA (((((..((((((((((((...((....))....--------))))))))))))....))))).(((...(((.(((.(.((((........))))...).))))))...)))....... ( -34.20) >DroYak_CAF1 7287 115 - 1 CCAAAUAGUUGACUCAGCCCGACCCGAGGGAUAUA----C-UAGCUGAGGCAACAGGUUUUGGCCCUGAAACGGGGCGCAUUUCAUUCAAAUGAAGGUGGGGCUUGUCAAAUGACGUACA ........(((((..(((((..((((((((...((----.-.((((..(....).)))).)).))))....)))).(((.((((((....)))))))))))))).))))).......... ( -35.40) >consensus CCAAAUAGUUGACUCAACUCGACCCGAGGGAUACA____CAUAGUUGAGUCAACAGGUUUUGGCCCUGAAACGGGGCGCAUUUUAUUCAAAUGAAGGUGGGGCUUGUCAAAUGACGUACA (((....((((((((((((...((....))............))))))))))))......((((((((...)))))).)).................))).(..((((....))))..). (-30.54 = -30.62 + 0.08)

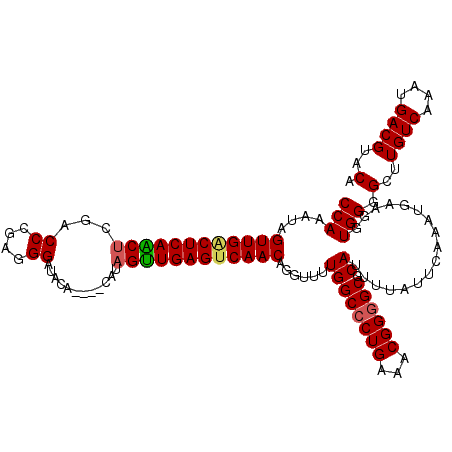
| Location | 8,167,271 – 8,167,388 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.18 |
| Mean single sequence MFE | -36.17 |
| Consensus MFE | -27.98 |
| Energy contribution | -28.33 |
| Covariance contribution | 0.35 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.77 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.508958 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8167271 117 - 22407834 CGGGCGGUCCUUCGGAUUACGACCUGACAUUAACAUGCUUCCAAAUAGUUGACUCAACUCGACCCGAGGGAUAG---GACAUAGUUGAGUCAACAGGUUUUGGCCCUGAAACGGGGCGCA ((((((((((...))))..)).)))).........(((...((((..((((((((((((....((........)---)....))))))))))))....))))((((((...))))))))) ( -40.70) >DroSec_CAF1 7147 116 - 1 CGGGCGGUCCUUCGGAUUACGACCUGACAUUAACAUGCUUCCAAAUAGUUGACUCAACUCGACCCGAGGGAUACA----CAUAGUUGAGUCAACAGGUUUUGGCCCUGAAACGGGGCGCA ((((((((((...))))..)).)))).........(((...((((..((((((((((((...((....)).....----...))))))))))))....))))((((((...))))))))) ( -38.60) >DroSim_CAF1 7664 120 - 1 CGGGCGGUCCUUCGGAUUACGACCUGACAUUAACAUGCUUCCAAAUAGUUGACUCAACUCGACCCGAGGGAUAUACAUACAUAGUUGAGUCAACAGGUUUUGGCCCUGAAACGGGGCACA ((((((((((...))))..)).))))...............((((..(((((((((((....((....)).(((......))))))))))))))....))))((((((...))))))... ( -36.40) >DroEre_CAF1 7660 112 - 1 CAGGCGGUCCCUAGGAUUACGACCUGACAUUAACAUGCUACCAAAUAGUUGGCUCAGCUCGACCCGAGGGAUAC--------AGUUGAGUCAACAGGUUUUGGCCCUGAAACGGAGCGCA ...(((.(((.((((..((.((((((..........((((.....))))((((((((((...((....))....--------)))))))))).)))))).))..))))....))).))). ( -39.30) >DroYak_CAF1 7327 115 - 1 CAGGCGGUCCUUAGGAUUACGACCUGACAUUAACAUGCUUCCAAAUAGUUGACUCAGCCCGACCCGAGGGAUAUA----C-UAGCUGAGGCAACAGGUUUUGGCCCUGAAACGGGGCGCA ((((((((((...))))..)).)))).........(((...((((..((((.((((((....((....)).....----.-..)))))).))))....))))((((((...))))))))) ( -35.70) >DroAna_CAF1 8684 94 - 1 CAGGAGGUCCUUAGGAUUACGACCUGACAUUAACAUGCCUGCAAAUAGUUGACGCAACAUGGCUUGAUGGAUGG--------CGUUGAGUCAACAGGUUGAG------------------ .....(((((...))))).(((((((.........(((...(((....)))..)))...(((((..(((.....--------)))..))))).)))))))..------------------ ( -26.30) >consensus CAGGCGGUCCUUAGGAUUACGACCUGACAUUAACAUGCUUCCAAAUAGUUGACUCAACUCGACCCGAGGGAUAC_____C_UAGUUGAGUCAACAGGUUUUGGCCCUGAAACGGGGCGCA ((((((((((...))))..)).)))).....................((((((((((((...((....))............))))))))))))........(((((.....)))))... (-27.98 = -28.33 + 0.35)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:37:51 2006