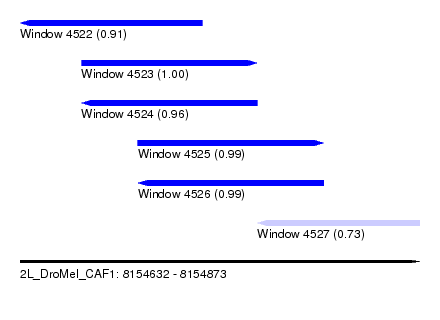
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,154,632 – 8,154,873 |
| Length | 241 |
| Max. P | 0.996638 |

| Location | 8,154,632 – 8,154,742 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 96.55 |
| Mean single sequence MFE | -26.10 |
| Consensus MFE | -21.98 |
| Energy contribution | -22.98 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.905559 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8154632 110 - 22407834 UAUUCCGCAUUCCGCAGAAUUUGAGUACAAAAUACUCAAAUUGCCAUUUUCGCUGUGUGGGUUAGUUUCGGCAUUAUUGUCUAGGCUUCCUUUCUACCAACAGAUUUGCC ......(((....(((..(((((((((.....))))))))))))........(((((((((..(((((.((((....)))).))))).....)))))..))))...))). ( -26.30) >DroSec_CAF1 20745 109 - 1 UAUUCCGCAUUCCGCAGAAUUUGAGUACAAAAUACUCAAAUUGCCAUU-UCGCUGUGUGGGUUAGUUUCGGCAUUAUUGUCUAGGCUUCCUUUCUACCAACAGAUUUGCC ......(((....(((..(((((((((.....))))))))))))....-...(((((((((..(((((.((((....)))).))))).....)))))..))))...))). ( -26.30) >DroSim_CAF1 22516 110 - 1 AAUUCCGCAUUCCGCAGAAUUUGAGUACAAAAUACUCAAAUUGCCAUUUUCGCUGUGUGGGUUAGUUUCGGCAUUAUUGUCUAGGCUUCCUUUCUACCAACAGAUUUGCC ......(((....(((..(((((((((.....))))))))))))........(((((((((..(((((.((((....)))).))))).....)))))..))))...))). ( -26.30) >DroEre_CAF1 20570 110 - 1 UAUUCCGCAUUCCGCAGAAUUUGAGUACAAAAUACUCAAUUUGCCAUUUUAGCUGUGUGGGUUAAUUUCGGCAUUUUUGUCUAGGCUUCCUUUCUACCAACAGAUUUGCC ...(((((((..((((((..(((((((.....)))))))))))).......)..)))))))........((((..(((((.((((.......))))...)))))..)))) ( -25.81) >DroYak_CAF1 20580 110 - 1 UAUUCCGCAUUCCGCAGAAUUUGAGUACAAAAUACUCAAUUCGCCAUUUUCGCUGUGUGGGUUAAUUUCGGCAGUUUUGUCUAGGCUUCCUUUCUACCAACAGAUUUGCC ...(((((((..((.((((((((((((.....)))))))......)))))))..)))))))........(((((.(((((.((((.......))))...))))).))))) ( -25.80) >consensus UAUUCCGCAUUCCGCAGAAUUUGAGUACAAAAUACUCAAAUUGCCAUUUUCGCUGUGUGGGUUAGUUUCGGCAUUAUUGUCUAGGCUUCCUUUCUACCAACAGAUUUGCC ......(((....(((..(((((((((.....))))))))))))........(((((((((..(((((.((((....)))).))))).....)))))..))))...))). (-21.98 = -22.98 + 1.00)


| Location | 8,154,669 – 8,154,775 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 92.65 |
| Mean single sequence MFE | -25.46 |
| Consensus MFE | -22.24 |
| Energy contribution | -22.84 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.73 |
| SVM RNA-class probability | 0.996638 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8154669 106 + 22407834 UGCCGAAACUAACCCACACAGCGAAAAUGGCAAUUUGAGUAUUUUGUACUCAAAUUCUGCGGAAUGCGGAAUA----AGAAA---UUAGCAUAAUUUCAAUUUCCUGUGCUUA ................(((((.((((........((((((((...))))))))(((((((.....))))))).----.((((---(((...)))))))..))))))))).... ( -25.90) >DroSec_CAF1 20782 105 + 1 UGCCGAAACUAACCCACACAGCGA-AAUGGCAAUUUGAGUAUUUUGUACUCAAAUUCUGCGGAAUGCGGAAUA----AGAAA---UUAGCAUAAUUUCAAUUUCCUGUGCUUA ................(((((.((-(((......((((((((...))))))))(((((((.....))))))).----.((((---(((...))))))).)))))))))).... ( -26.90) >DroSim_CAF1 22553 106 + 1 UGCCGAAACUAACCCACACAGCGAAAAUGGCAAUUUGAGUAUUUUGUACUCAAAUUCUGCGGAAUGCGGAAUU----AGAAA---UUAGCAUAAUUUCAAUUUCCUGUGCUUA ................(((((.((((.........(((((((...)))))))((((((((.....))))))))----.((((---(((...)))))))..))))))))).... ( -25.90) >DroEre_CAF1 20607 104 + 1 UGCCGAAAUUAACCCACACAGCUAAAAUGGCAAAUUGAGUAUUUUGUACUCAAAUUCUGCGGAAUGCGGAAUA----AUAAA---UUUGCAUAAUUUCAAUUUCCUGUGCU-- ....(((((((.......((.......))(((((((((((((...))))))..(((((((.....))))))).----...))---))))).))))))).............-- ( -23.70) >DroYak_CAF1 20617 113 + 1 UGCCGAAAUUAACCCACACAGCGAAAAUGGCGAAUUGAGUAUUUUGUACUCAAAUUCUGCGGAAUGCGGAAUACGGAAUAAAUCAUUUGCAUAAUUUCAACUUCCUGUGCUUA ................(((((.((((((.(((((((((((((...))))))))(((((((.....))))))).............)))))...))).....)))))))).... ( -24.90) >consensus UGCCGAAACUAACCCACACAGCGAAAAUGGCAAUUUGAGUAUUUUGUACUCAAAUUCUGCGGAAUGCGGAAUA____AGAAA___UUAGCAUAAUUUCAAUUUCCUGUGCUUA ................(((((.((((.(((....((((((((...))))))))(((((((.....)))))))........................))).))))))))).... (-22.24 = -22.84 + 0.60)

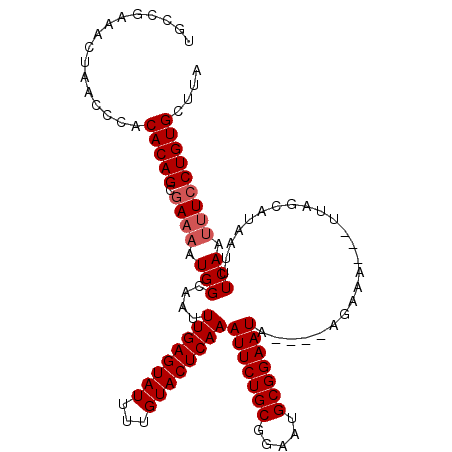
| Location | 8,154,669 – 8,154,775 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 92.65 |
| Mean single sequence MFE | -25.34 |
| Consensus MFE | -22.32 |
| Energy contribution | -22.52 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.956487 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8154669 106 - 22407834 UAAGCACAGGAAAUUGAAAUUAUGCUAA---UUUCU----UAUUCCGCAUUCCGCAGAAUUUGAGUACAAAAUACUCAAAUUGCCAUUUUCGCUGUGUGGGUUAGUUUCGGCA ...((...((((...(((((((...)))---)))).----..))))....((((((.((((((((((.....))))))))))((.......))..)))))).........)). ( -25.80) >DroSec_CAF1 20782 105 - 1 UAAGCACAGGAAAUUGAAAUUAUGCUAA---UUUCU----UAUUCCGCAUUCCGCAGAAUUUGAGUACAAAAUACUCAAAUUGCCAUU-UCGCUGUGUGGGUUAGUUUCGGCA ...((...((((...(((((((...)))---)))).----..))))....((((((.((((((((((.....))))))))))((....-..))..)))))).........)). ( -26.30) >DroSim_CAF1 22553 106 - 1 UAAGCACAGGAAAUUGAAAUUAUGCUAA---UUUCU----AAUUCCGCAUUCCGCAGAAUUUGAGUACAAAAUACUCAAAUUGCCAUUUUCGCUGUGUGGGUUAGUUUCGGCA ...((...((((...(((((((...)))---)))).----..))))....((((((.((((((((((.....))))))))))((.......))..)))))).........)). ( -25.80) >DroEre_CAF1 20607 104 - 1 --AGCACAGGAAAUUGAAAUUAUGCAAA---UUUAU----UAUUCCGCAUUCCGCAGAAUUUGAGUACAAAAUACUCAAUUUGCCAUUUUAGCUGUGUGGGUUAAUUUCGGCA --.((((((....(((((((...(((((---.....----.((((.((.....)).))))(((((((.....)))))))))))).)))))))))))))............... ( -21.80) >DroYak_CAF1 20617 113 - 1 UAAGCACAGGAAGUUGAAAUUAUGCAAAUGAUUUAUUCCGUAUUCCGCAUUCCGCAGAAUUUGAGUACAAAAUACUCAAUUCGCCAUUUUCGCUGUGUGGGUUAAUUUCGGCA ...((...((((....(((((((....))))))).))))....(((((((..((.((((((((((((.....)))))))......)))))))..))))))).........)). ( -27.00) >consensus UAAGCACAGGAAAUUGAAAUUAUGCUAA___UUUCU____UAUUCCGCAUUCCGCAGAAUUUGAGUACAAAAUACUCAAAUUGCCAUUUUCGCUGUGUGGGUUAGUUUCGGCA ...((....((((((((....((((.....................))))((((((.((((((((((.....))))))))))((.......))..)))))))))))))).)). (-22.32 = -22.52 + 0.20)

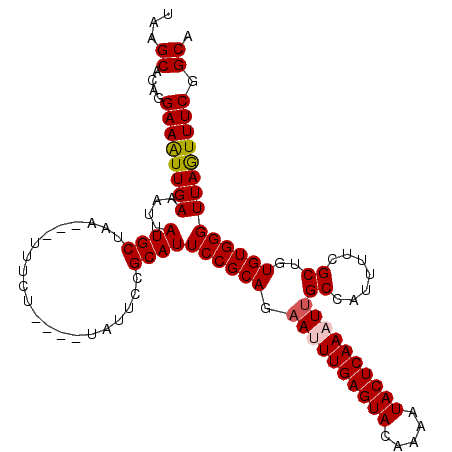
| Location | 8,154,703 – 8,154,815 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 89.63 |
| Mean single sequence MFE | -30.14 |
| Consensus MFE | -25.32 |
| Energy contribution | -25.16 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.43 |
| SVM RNA-class probability | 0.993862 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8154703 112 + 22407834 UUGAGUAUUUUGUACUCAAAUUCUGCGGAAUGCGGAAUA----AGAAA---UUAGCAUAAUUUCAAUUUCCUGUGCUUAAAGUUUUGGGAAACCCAGAAGCAAGGGUACUUUUUUAUAG ((((((((...))))))))(((((((.....))))))).----.((((---(((...)))))))........((((((...(((((((....))..)))))..)))))).......... ( -28.60) >DroSec_CAF1 20815 111 + 1 UUGAGUAUUUUGUACUCAAAUUCUGCGGAAUGCGGAAUA----AGAAA---UUAGCAUAAUUUCAAUUUCCUGUGCUUAAGGUUUUGGGAAACCCAGAAGCAAGGGUACUU-UUUAUGG ((((((((...))))))))(((((((.....))))))).----.((((---(((...)))))))........((((((..(.(((((((...))))))).)..))))))..-....... ( -31.30) >DroSim_CAF1 22587 112 + 1 UUGAGUAUUUUGUACUCAAAUUCUGCGGAAUGCGGAAUU----AGAAA---UUAGCAUAAUUUCAAUUUCCUGUGCUUAAGGUUUUGGGAAACCCAGAAGCAAGGGUACUUUUUUAUAG .(((((((...)))))))((((((((.....))))))))----.((((---(((...)))))))........((((((..(.(((((((...))))))).)..)))))).......... ( -31.50) >DroEre_CAF1 20641 109 + 1 UUGAGUAUUUUGUACUCAAAUUCUGCGGAAUGCGGAAUA----AUAAA---UUUGCAUAAUUUCAAUUUCCUGUGCU---GGGUUUGGGAAACCCAAAAGCAAGGGUACUUUUGUUGUU ((((((((...))))))))(((((((.....))))))).----.....---.......(((..(((...(((.((((---...((.((....)).)).)))).))).....)))..))) ( -28.20) >DroYak_CAF1 20651 119 + 1 UUGAGUAUUUUGUACUCAAAUUCUGCGGAAUGCGGAAUACGGAAUAAAUCAUUUGCAUAAUUUCAACUUCCUGUGCUUAAGAUUUUGGGAAACCCAGAAGCAAGGGUAAUCUUGCAGAG ((((((((...))))))))(((((((.....))))))).............((((((..(((......((((.((((......((((((...)))))))))))))).)))..)))))). ( -31.10) >consensus UUGAGUAUUUUGUACUCAAAUUCUGCGGAAUGCGGAAUA____AGAAA___UUAGCAUAAUUUCAAUUUCCUGUGCUUAAGGUUUUGGGAAACCCAGAAGCAAGGGUACUUUUUUAUAG ((((((((...))))))))(((((((.....)))))))...............................(((.((((......((((((...)))))))))).)))............. (-25.32 = -25.16 + -0.16)

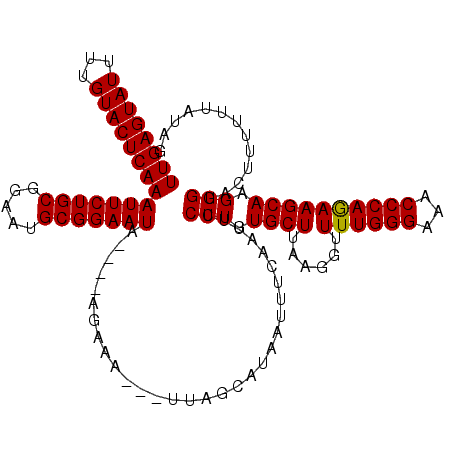

| Location | 8,154,703 – 8,154,815 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 89.63 |
| Mean single sequence MFE | -24.10 |
| Consensus MFE | -18.80 |
| Energy contribution | -19.08 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.78 |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.988357 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8154703 112 - 22407834 CUAUAAAAAAGUACCCUUGCUUCUGGGUUUCCCAAAACUUUAAGCACAGGAAAUUGAAAUUAUGCUAA---UUUCU----UAUUCCGCAUUCCGCAGAAUUUGAGUACAAAAUACUCAA ..............((((((((..((((((....)))))).))))).))).....(((((((...)))---)))).----.((((.((.....)).))))(((((((.....))))))) ( -21.60) >DroSec_CAF1 20815 111 - 1 CCAUAAA-AAGUACCCUUGCUUCUGGGUUUCCCAAAACCUUAAGCACAGGAAAUUGAAAUUAUGCUAA---UUUCU----UAUUCCGCAUUCCGCAGAAUUUGAGUACAAAAUACUCAA .......-......((((((((..((((((....)))))).))))).))).....(((((((...)))---)))).----.((((.((.....)).))))(((((((.....))))))) ( -24.40) >DroSim_CAF1 22587 112 - 1 CUAUAAAAAAGUACCCUUGCUUCUGGGUUUCCCAAAACCUUAAGCACAGGAAAUUGAAAUUAUGCUAA---UUUCU----AAUUCCGCAUUCCGCAGAAUUUGAGUACAAAAUACUCAA ..............((((((((..((((((....)))))).))))).))).....(((((((...)))---)))).----(((((.((.....)).)))))((((((.....)))))). ( -25.20) >DroEre_CAF1 20641 109 - 1 AACAACAAAAGUACCCUUGCUUUUGGGUUUCCCAAACCC---AGCACAGGAAAUUGAAAUUAUGCAAA---UUUAU----UAUUCCGCAUUCCGCAGAAUUUGAGUACAAAAUACUCAA ......(((((((....)))))))(((((.....)))))---......((((....(((((.....))---)))..----..))))((.....)).....(((((((.....))))))) ( -20.60) >DroYak_CAF1 20651 119 - 1 CUCUGCAAGAUUACCCUUGCUUCUGGGUUUCCCAAAAUCUUAAGCACAGGAAGUUGAAAUUAUGCAAAUGAUUUAUUCCGUAUUCCGCAUUCCGCAGAAUUUGAGUACAAAAUACUCAA .(((((((((((((((........)))).......))))))..((...((((....(((((((....))))))).))))(.....))).....)))))..(((((((.....))))))) ( -28.71) >consensus CUAUAAAAAAGUACCCUUGCUUCUGGGUUUCCCAAAACCUUAAGCACAGGAAAUUGAAAUUAUGCUAA___UUUCU____UAUUCCGCAUUCCGCAGAAUUUGAGUACAAAAUACUCAA ..............((((((((..((((((....)))))).))))).)))...............................((((.((.....)).))))(((((((.....))))))) (-18.80 = -19.08 + 0.28)



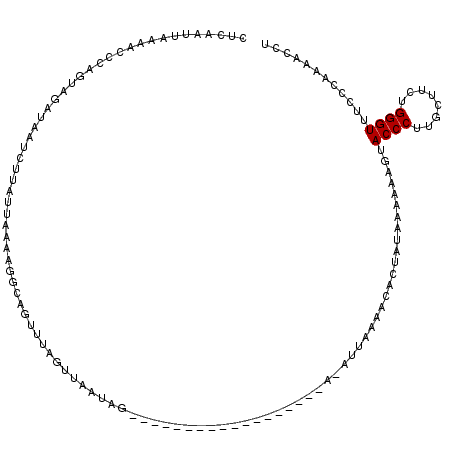
| Location | 8,154,775 – 8,154,873 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 69.79 |
| Mean single sequence MFE | -16.94 |
| Consensus MFE | -5.00 |
| Energy contribution | -5.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.30 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.725895 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8154775 98 - 22407834 CUCAAUUAAAACCCAGUUGAUAAUCCUAUUAAAAGGCAGUUUAGUUAAUAG------------------A-A---AAACACUAUAAAAAAGUACCCUUGCUUCUGGGUUUCCCAAAACUU ........((((((((((((((....)))))).((((((....(((.....------------------.-.---.)))(((.......)))....)))))))))))))).......... ( -15.40) >DroSec_CAF1 20887 100 - 1 CUCAAUUAAAACCCAGUAGAUAAUCUUAUUAAAAGGCAGUUAAGAUAAUAG------------------A-AUUAAAACACCAUAAA-AAGUACCCUUGCUUCUGGGUUUCCCAAAACCU ........((((((((......((((((............)))))).....------------------.-................-(((((....))))))))))))).......... ( -14.50) >DroSim_CAF1 22659 101 - 1 CUCAAUUAAAACCCAGUAGAUAAUCUUAUUAAAAGGUAGUUUAGUUAAUAG------------------A-AUUAAAACACUAUAAAAAAGUACCCUUGCUUCUGGGUUUCCCAAAACCU ........((((((((.((.(((...((((.....(((((((((((.....------------------)-)))))...))))).....))))...))))).)))))))).......... ( -16.30) >DroEre_CAF1 20711 119 - 1 CUCAGUAGGAACACAUAAGUUGAACUUAUUAUAAAGCUGUUCAGUUAAUAAGCCCUAGACAGAAUGGACACAUUAAGACCAACAACAAAAGUACCCUUGCUUUUGGGUUUCCCAAACCC- .......(((((......((((..(((.........((((..((..........))..))))((((....)))))))..))))..((((((((....)))))))).)))))........- ( -21.30) >DroYak_CAF1 20730 85 - 1 -----------------ACUGAAACUUAUUGUAAAACAGUUUAGUUAAUAA------------------ACAUUAAAACCCUCUGCAAGAUUACCCUUGCUUCUGGGUUUCCCAAAAUCU -----------------.....((((.((((.....))))..)))).....------------------......((((((...(((((......)))))....)))))).......... ( -17.20) >consensus CUCAAUUAAAACCCAGUAGAUAAUCUUAUUAAAAGGCAGUUUAGUUAAUAG__________________A_AUUAAAACACUAUAAAAAAGUACCCUUGCUUCUGGGUUUCCCAAAACCU ............................................................................................((((........))))............ ( -5.00 = -5.00 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:37:46 2006