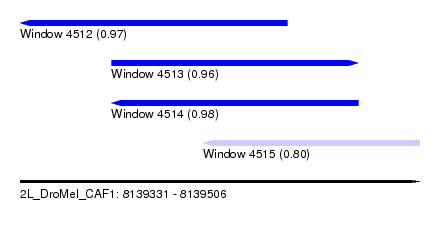
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,139,331 – 8,139,506 |
| Length | 175 |
| Max. P | 0.983602 |

| Location | 8,139,331 – 8,139,448 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.53 |
| Mean single sequence MFE | -33.54 |
| Consensus MFE | -20.79 |
| Energy contribution | -21.87 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.973988 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8139331 117 - 22407834 --C-AAUCCUCCUCCAACAUCGACACUUGGAGGAUUCUUUACCACAGGUGAAGUUGCACCUGUUGUUGCGCCAUUUGGAACACAAGCGUCACCAGCAACACAAUCUGCAGGAUUCUUUAA --.-(((((((((((((.........))))))))..(((((((...))))))).((((..(((((((((((...(((.....))))))....)))))))).....)))))))))...... ( -34.10) >DroSec_CAF1 4737 117 - 1 --C-AAUCCUCCUCCAACAUCGGCACUAGGAGGAUUCUUUACCACAGGUGAAGUUGCACCUGUUGUUGGGCCAUUUGCAACACAAGCGACACCAGCAACACAAUCUGCAGGAUUCUUUAA --.-(((((((((((......))....)))))))))(((((((...)))))))(((((..(((((((((.....((((.......))))..))))))))).....))))).......... ( -36.40) >DroSim_CAF1 5730 117 - 1 --C-AAUCCUCCUCCAACAUCGGCACUAGGAGGAUUCUUUACCACAGGUGAAGUUGCACCUGUUGUUGGGCCAUUUGCAACACAAGCGACACCAGCAACACAAUCUGCAGGAUUCUUUAA --.-(((((((((((......))....)))))))))(((((((...)))))))(((((..(((((((((.....((((.......))))..))))))))).....))))).......... ( -36.40) >DroEre_CAF1 4925 117 - 1 --C-AAUCCUCCCCCAACAUCGGUACUAGGAGGAUUCUUUACCACAGGUGAAGUUCCACCUGCUGUUGGACCAUUUGCAACACAAGAGACAGCAGCAACGCAAUCUGUAGGAUUCUUAAA --.-((((((((.((......)).....)))))))).....((((((((...(((....((((((((......((((.....)))).)))))))).)))...)))))).))......... ( -31.30) >DroYak_CAF1 4705 105 - 1 --C-AAUCCUCCCCCCACAUCGACACUAGGAGGAUUCUUUACCACAGGUGAAGUUGCACCUGUUGUUGGA------------CAAGCGACUCCAGCAACACAACCUGUAGGAUUCUUAAA --.-((((((((................)))))))).....((((((((...(((((....((((((...------------..))))))....)))))...)))))).))......... ( -33.69) >DroAna_CAF1 4789 106 - 1 UCCAAGCCUUCCCCCAGCAGCAGUGCCCGGAAAUUUCUUCACCCCCGG---AGUUGUCCCAGCUGUUGCUCCAACUGCAACACCAGCGCCAGCAGCA---------GGAGGAUUUUUG-- ..((((...(((.(((((((((((....(((.....((((......))---))...)))..)))))))))....((((.............))))..---------)).)))..))))-- ( -29.32) >consensus __C_AAUCCUCCCCCAACAUCGGCACUAGGAGGAUUCUUUACCACAGGUGAAGUUGCACCUGUUGUUGGGCCAUUUGCAACACAAGCGACACCAGCAACACAAUCUGCAGGAUUCUUUAA ....((((((((................)))))))).....((..(((((......)))))((((((((......................))))))))..........))......... (-20.79 = -21.87 + 1.08)


| Location | 8,139,371 – 8,139,479 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.81 |
| Mean single sequence MFE | -37.22 |
| Consensus MFE | -23.98 |
| Energy contribution | -24.18 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.963228 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8139371 108 + 22407834 UUCCAAAUGGCGCAACAACAGGUGCAACUUCACCUGUGGUAAAGAAUCCUCCAAGUGUCGAUGUUGGAGGAGGAUU-G--------CCU---GGUGGAACUGUUAAAGGUAUUGUUGGAA .(((((...((((........))))...((((((...(((((....((((((((.........))))))))...))-)--------)).---))))))................))))). ( -34.40) >DroSec_CAF1 4777 108 + 1 UUGCAAAUGGCCCAACAACAGGUGCAACUUCACCUGUGGUAAAGAAUCCUCCUAGUGCCGAUGUUGGAGGAGGAUU-G--------CCU---GGUGGUACUGUUAAAGGUAUUGUUGGAA ..((.....(((.....(((((((......))))))))))....(((((((((...((....))...)))))))))-)--------)(.---.(..(((((......)))))..)..).. ( -36.40) >DroSim_CAF1 5770 108 + 1 UUGCAAAUGGCCCAACAACAGGUGCAACUUCACCUGUGGUAAAGAAUCCUCCUAGUGCCGAUGUUGGAGGAGGAUU-G--------CCU---GGUGGUACUGUUAAAGGUAUUGUUGGAA ..((.....(((.....(((((((......))))))))))....(((((((((...((....))...)))))))))-)--------)(.---.(..(((((......)))))..)..).. ( -36.40) >DroEre_CAF1 4965 108 + 1 UUGCAAAUGGUCCAACAGCAGGUGGAACUUCACCUGUGGUAAAGAAUCCUCCUAGUACCGAUGUUGGGGGAGGAUU-G--------CCU---GGUGGAACUGUUAAAGGUAUUGUUGGAA ..........(((((((((((((((....)))))))).......(((((((((...((....))...)))))))))-(--------(((---..............))))..))))))). ( -36.84) >DroYak_CAF1 4743 98 + 1 ----------UCCAACAACAGGUGCAACUUCACCUGUGGUAAAGAAUCCUCCUAGUGUCGAUGUGGGGGGAGGAUU-G--------CCU---GGUGGAACUGUUAAAGGUAUUGUUGGAA ----------(((((((((....(((..((((((...(((((....((((((((.........))))))))...))-)--------)).---))))))..))).....))..))))))). ( -35.30) >DroAna_CAF1 4818 117 + 1 UUGCAGUUGGAGCAACAGCUGGGACAACU---CCGGGGGUGAAGAAAUUUCCGGGCACUGCUGCUGGGGGAAGGCUUGGAGGAGGUCCUGCAGAGGGAGCGGCCAAAGGUGGAGUAGCAA ((.((((((......)))))).))..(((---((((..((......))..))((.(.((.((.(((.((((...((....))...)))).))).)).)).).))......)))))..... ( -44.00) >consensus UUGCAAAUGGCCCAACAACAGGUGCAACUUCACCUGUGGUAAAGAAUCCUCCUAGUGCCGAUGUUGGAGGAGGAUU_G________CCU___GGUGGAACUGUUAAAGGUAUUGUUGGAA ...........(((((((((((((......)))))))(((....(((((((((....((......)))))))))))..........((.......)).)))...........)))))).. (-23.98 = -24.18 + 0.20)


| Location | 8,139,371 – 8,139,479 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.81 |
| Mean single sequence MFE | -30.68 |
| Consensus MFE | -20.78 |
| Energy contribution | -21.47 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.22 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.95 |
| SVM RNA-class probability | 0.983602 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8139371 108 - 22407834 UUCCAACAAUACCUUUAACAGUUCCACC---AGG--------C-AAUCCUCCUCCAACAUCGACACUUGGAGGAUUCUUUACCACAGGUGAAGUUGCACCUGUUGUUGCGCCAUUUGGAA .....................(((((..---.((--------(-.....((((((((.........))))))))......((.(((((((......))))))).))...)))...))))) ( -31.10) >DroSec_CAF1 4777 108 - 1 UUCCAACAAUACCUUUAACAGUACCACC---AGG--------C-AAUCCUCCUCCAACAUCGGCACUAGGAGGAUUCUUUACCACAGGUGAAGUUGCACCUGUUGUUGGGCCAUUUGCAA .(((((((....................---(((--------.-(((((((((((......))....))))))))))))....(((((((......)))))))))))))).......... ( -32.10) >DroSim_CAF1 5770 108 - 1 UUCCAACAAUACCUUUAACAGUACCACC---AGG--------C-AAUCCUCCUCCAACAUCGGCACUAGGAGGAUUCUUUACCACAGGUGAAGUUGCACCUGUUGUUGGGCCAUUUGCAA .(((((((....................---(((--------.-(((((((((((......))....))))))))))))....(((((((......)))))))))))))).......... ( -32.10) >DroEre_CAF1 4965 108 - 1 UUCCAACAAUACCUUUAACAGUUCCACC---AGG--------C-AAUCCUCCCCCAACAUCGGUACUAGGAGGAUUCUUUACCACAGGUGAAGUUCCACCUGCUGUUGGACCAUUUGCAA .(((((((....................---(((--------.-((((((((.((......)).....))))))))))).....((((((......)))))).))))))).......... ( -29.90) >DroYak_CAF1 4743 98 - 1 UUCCAACAAUACCUUUAACAGUUCCACC---AGG--------C-AAUCCUCCCCCCACAUCGACACUAGGAGGAUUCUUUACCACAGGUGAAGUUGCACCUGUUGUUGGA---------- .(((((((....................---(((--------.-((((((((................)))))))))))....(((((((......))))))))))))))---------- ( -29.09) >DroAna_CAF1 4818 117 - 1 UUGCUACUCCACCUUUGGCCGCUCCCUCUGCAGGACCUCCUCCAAGCCUUCCCCCAGCAGCAGUGCCCGGAAAUUUCUUCACCCCCGG---AGUUGUCCCAGCUGUUGCUCCAACUGCAA .(((............(((.((.......)).(((.....)))..))).......(((((((((....(((.....((((......))---))...)))..)))))))))......))). ( -29.80) >consensus UUCCAACAAUACCUUUAACAGUUCCACC___AGG________C_AAUCCUCCCCCAACAUCGGCACUAGGAGGAUUCUUUACCACAGGUGAAGUUGCACCUGUUGUUGGGCCAUUUGCAA .(((((((....................................((((((((................))))))))........((((((......)))))).))))))).......... (-20.78 = -21.47 + 0.70)

| Location | 8,139,411 – 8,139,506 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.90 |
| Mean single sequence MFE | -19.96 |
| Consensus MFE | -12.70 |
| Energy contribution | -11.93 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.802597 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8139411 95 - 22407834 -CAG--------UUCCAUUUG---UUGACA-CACAAGCUGUUCCAACAAUACCUUUAACAGUUCCACC---AGG--------C-AAUCCUCCUCCAACAUCGACACUUGGAGGAUUCUUU -...--------......(((---(((..(-((.....)))..))))))...................---(((--------.-((((((((................))))))))))). ( -17.29) >DroSec_CAF1 4817 95 - 1 -CAG--------UUCCAUUUG---UUGACA-CACAAGCUGUUCCAACAAUACCUUUAACAGUACCACC---AGG--------C-AAUCCUCCUCCAACAUCGGCACUAGGAGGAUUCUUU -...--------......(((---(((..(-((.....)))..))))))...................---(((--------.-(((((((((((......))....)))))))))))). ( -19.70) >DroSim_CAF1 5810 95 - 1 -CAG--------UUCCAUUUG---UUGACA-CACAAGCUGUUCCAACAAUACCUUUAACAGUACCACC---AGG--------C-AAUCCUCCUCCAACAUCGGCACUAGGAGGAUUCUUU -...--------......(((---(((..(-((.....)))..))))))...................---(((--------.-(((((((((((......))....)))))))))))). ( -19.70) >DroEre_CAF1 5005 95 - 1 -CAG--------UUCCAUUUG---UUGACA-CACAAGCUGUUCCAACAAUACCUUUAACAGUUCCACC---AGG--------C-AAUCCUCCCCCAACAUCGGUACUAGGAGGAUUCUUU -...--------......(((---(((..(-((.....)))..))))))...................---(((--------.-((((((((.((......)).....))))))))))). ( -17.70) >DroYak_CAF1 4773 95 - 1 -CAG--------UUCCAUUUG---UUGACA-CACAAGCUGUUCCAACAAUACCUUUAACAGUUCCACC---AGG--------C-AAUCCUCCCCCCACAUCGACACUAGGAGGAUUCUUU -...--------......(((---(((..(-((.....)))..))))))...................---(((--------.-((((((((................))))))))))). ( -16.29) >DroAna_CAF1 4855 120 - 1 GCAGCAGGCUUCUUCAAUCCGGGAUUAACUCCGCCAGCGGUUGCUACUCCACCUUUGGCCGCUCCCUCUGCAGGACCUCCUCCAAGCCUUCCCCCAGCAGCAGUGCCCGGAAAUUUCUUC ((.....))........((((((.........(((((.(((.(.....).))).)))))......(.((((.(((.....)))..((.........)).)))).)))))))......... ( -29.10) >consensus _CAG________UUCCAUUUG___UUGACA_CACAAGCUGUUCCAACAAUACCUUUAACAGUUCCACC___AGG________C_AAUCCUCCCCCAACAUCGGCACUAGGAGGAUUCUUU ....................................((((((..............))))))......................((((((((................)))))))).... (-12.70 = -11.93 + -0.77)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:37:35 2006