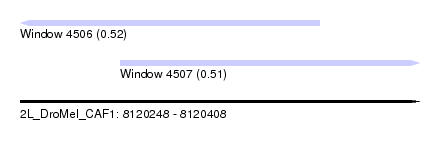
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,120,248 – 8,120,408 |
| Length | 160 |
| Max. P | 0.515546 |

| Location | 8,120,248 – 8,120,368 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.06 |
| Mean single sequence MFE | -38.07 |
| Consensus MFE | -24.36 |
| Energy contribution | -24.75 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.64 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.515546 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
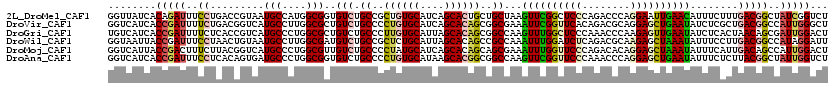
>2L_DroMel_CAF1 8120248 120 - 22407834 GGUUAUCACAGAUUUCCUGACCGUAAUGCCAUGGCGGUGUCUGCCGCUGUGCAUCAGCACUGCUGCUAAGUUCGGCUCCCAGACCCAGGAAUUGAACAUUUCUUUGACGGCUAUCGGUCU ((..(((...)))..)).(((((....(((.(((((((....)))((.((((....)))).)).)))).((((((.(((........))).))))))...........)))...))))). ( -40.20) >DroVir_CAF1 85675 120 - 1 GGUCAUCACCGAUUUUCUGACGGUCAUGCCUUGGCGCUGUCUGCCCCUGUGCAUCAGCACAGCGGCGAAAUUCGGUUCACAGACGCAGGAGCUGAAUAUCUCGCUGACGGCCAUUGGGCU .((((..(((((.((((.((((((...((....)))))))).(((.((((((....)))))).))))))).)))))........((..((........))..))))))((((....)))) ( -46.90) >DroGri_CAF1 78712 120 - 1 UGUCAUCACCGAUUUUCUCACCGUCAUGCCCUGGCGCUGUCUGCCCUUGUGCAUUAGCACAGCGGCCAAGUUUGGCUCCCAAACCCAAGAGUUGAAUAUCUCACUAACAGCGAUUGGACU ........((((((........((((.....))))(((((..(((.((((((....)))))).)))...(((..((((..........))))..))).........)))))))))))... ( -35.10) >DroWil_CAF1 93029 120 - 1 GGUAAUUACCGAUUUCCUAACUGUAAUGCCUUGGCGAUGUCUGCCGCUCUGCAUUAGCACAGCCGCCAAAUUUGGAUCUCAGACGCAAGAGCUAAAUAUUUCCUUGACGGCCAUAGGAUU (((....)))....(((((..((((((((...((((........))))..))))).)))..((((.(((((((((.(((........))).))))))......))).))))..))))).. ( -29.00) >DroMoj_CAF1 74978 120 - 1 GGUCAUUACCGACUUUCUUACGGUCAUGCCCUGGCGUUGUCUGCCCCUAUGCAUCAGCACAGCAGCGAAAUUUGGUUCCCAGACACAGGAGCUAAAUAUUUCAUUGACAGCCAUUGGACU ((.(((.((((.........)))).))).))((((..((((.((..((.(((....))).))..))(((((((((((((........))))))))..)))))...))))))))....... ( -34.80) >DroAna_CAF1 79102 120 - 1 GGUCAUCACCGAUUUCCUCACAGUGAUGCCCUGGCGGUGUCUGCCCCUGUGCAUAAGCACGGCGGCCAAGUUCGGUUCCCAAACCCAGGAGCUGAAUAUUUCUCUUACGGCUAUUGGUCU ((.((((((.((.....))...)))))).))((((.(((...(((.((((((....)))))).)))...((((((((((........))))))))))........))).))))....... ( -42.40) >consensus GGUCAUCACCGAUUUCCUCACCGUCAUGCCCUGGCGCUGUCUGCCCCUGUGCAUCAGCACAGCGGCCAAAUUCGGUUCCCAGACCCAGGAGCUGAAUAUUUCACUGACGGCCAUUGGACU ........(((((..((.........(((....)))..(((.((..((((((....))))))..))...((((((((((........))))))))))........)))))..)))))... (-24.36 = -24.75 + 0.39)
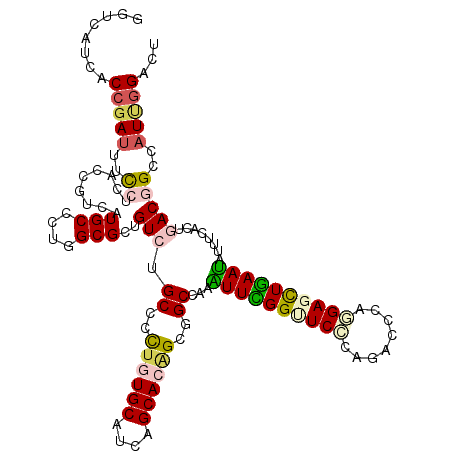
| Location | 8,120,288 – 8,120,408 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.94 |
| Mean single sequence MFE | -45.15 |
| Consensus MFE | -29.37 |
| Energy contribution | -30.77 |
| Covariance contribution | 1.39 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.65 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.508506 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8120288 120 + 22407834 GGGAGCCGAACUUAGCAGCAGUGCUGAUGCACAGCGGCAGACACCGCCAUGGCAUUACGGUCAGGAAAUCUGUGAUAACCAACUGCAGGCAUUGGAAUGCUGUGCGAAUUAACGGCUCGC ..((((((...((((((....))))))((((((((((((((.....((.((((......))))))...))))).....((((.((....))))))..)))))))))......)))))).. ( -46.30) >DroVir_CAF1 85715 120 + 1 GUGAACCGAAUUUCGCCGCUGUGCUGAUGCACAGGGGCAGACAGCGCCAAGGCAUGACCGUCAGAAAAUCGGUGAUGACCAGCUGCAGGCACUGGAAUGCGGUGCGUAUCAGCGGCUCGC ((((..........(((.((((((....)))))).))).......(((..(((((.((((.........)))).))).)).((((.(.((((((.....)))))).)..))))))))))) ( -52.30) >DroGri_CAF1 78752 120 + 1 GGGAGCCAAACUUGGCCGCUGUGCUAAUGCACAAGGGCAGACAGCGCCAGGGCAUGACGGUGAGAAAAUCGGUGAUGACAAGCUGCAGGCACUGGAAUGCGGUGCGUAUCAGCGGCUCAC ..(((((...((((((.(((((.....(((......))).))))))))))).(((.(((((......))).)).)))....((((.(.((((((.....)))))).)..))))))))).. ( -47.90) >DroWil_CAF1 93069 120 + 1 GAGAUCCAAAUUUGGCGGCUGUGCUAAUGCAGAGCGGCAGACAUCGCCAAGGCAUUACAGUUAGGAAAUCGGUAAUUACCAAUUGCAGGCAUUGGAAGGCAGUGCGUAUCAAUGGCUCGC ((..(((....(((((((((.(((....))).)))(.....)..))))))(((......))).)))..))(((....)))....((((.((((((.(.((...)).).)))))).)).)) ( -35.90) >DroMoj_CAF1 75018 120 + 1 GGGAACCAAAUUUCGCUGCUGUGCUGAUGCAUAGGGGCAGACAACGCCAGGGCAUGACCGUAAGAAAGUCGGUAAUGACCAGCUGCAGGCAUUGAAAGGCAGUGCGUAUCAGCGGCUCUC ((((..........(((.((((((....)))))).))).......(((.((.(((.((((.........)))).))).)).((((.(.((((((.....)))))).)..))))))))))) ( -44.40) >DroAna_CAF1 79142 120 + 1 GGGAACCGAACUUGGCCGCCGUGCUUAUGCACAGGGGCAGACACCGCCAGGGCAUCACUGUGAGGAAAUCGGUGAUGACCAACUGCAAGCACUGGAAGGCUGUCCGGAUAAGUGGCUCGC .............((((((.((((....))))..((((((.(....((((((((((((((.........)))))))).))...((....))))))...)))))))......))))))... ( -44.10) >consensus GGGAACCAAACUUGGCCGCUGUGCUGAUGCACAGGGGCAGACACCGCCAGGGCAUGACCGUCAGAAAAUCGGUGAUGACCAACUGCAGGCACUGGAAGGCAGUGCGUAUCAGCGGCUCGC ..............(((.((((((....)))))).))).......(((..(((((.((((.........)))).))).)).((((...((((((.....))))))....))))))).... (-29.37 = -30.77 + 1.39)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:37:27 2006