
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,115,906 – 8,116,038 |
| Length | 132 |
| Max. P | 0.995022 |

| Location | 8,115,906 – 8,115,998 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 92.98 |
| Mean single sequence MFE | -9.20 |
| Consensus MFE | -8.80 |
| Energy contribution | -8.80 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.35 |
| SVM RNA-class probability | 0.992761 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8115906 92 + 22407834 AGUUGAAAAGCAACUUUCAGCUAAUAUAGAUUAAUAACAAAUAAAGACCGAAAAACCACAAAAA---AAA--AACAAAUAUUUUAAAGCAAGCGCUU ((((((((......))))))))..........................................---...--.............((((....)))) ( -8.90) >DroSec_CAF1 71846 95 + 1 AGUUGAAACGCAACUUUCAGCUAAUAUAGAUUAAUAACAAAUAAAGACCGAAAAACCACAAAAAAAAAAA--AACAAAUAUUUUAAAGCAAGCGCUU ((((((((......))))))))................................................--.............((((....)))) ( -8.60) >DroSim_CAF1 71942 94 + 1 AGUUGAAAAGCAACUUUCAGCUAAUAUAGAUUAAUAACAAAUAAAGACCGAAAAACCACAAAAAA-AAAA--AACAAAUAUUUUAAAGCAAGCGCUU ((((((((......))))))))...........................................-....--.............((((....)))) ( -8.90) >DroEre_CAF1 72527 86 + 1 AGUUGAAAUGCAAAUUUCAGCUAAUUUAGAUUAAUAACAAAUAAAGACCGAAAAACCACAAAAAA-----------AAUAUUUUAAAGCAAGCGCUU (((((((((....)))))))))...........................................-----------.........((((....)))) ( -10.70) >DroYak_CAF1 73759 97 + 1 AGUUGAAAAGCAACUUUCAGCUAAUAUAGAUUAAUAACAAAUAAAGACCGAAAAACCACAAAAAAAAAAAUCAACAAAUAUUUUAAAGCAAGCGCUU ((((((((......))))))))...............................................................((((....)))) ( -8.90) >consensus AGUUGAAAAGCAACUUUCAGCUAAUAUAGAUUAAUAACAAAUAAAGACCGAAAAACCACAAAAAA_AAAA__AACAAAUAUUUUAAAGCAAGCGCUU ((((((((......))))))))...............................................................((((....)))) ( -8.80 = -8.80 + -0.00)

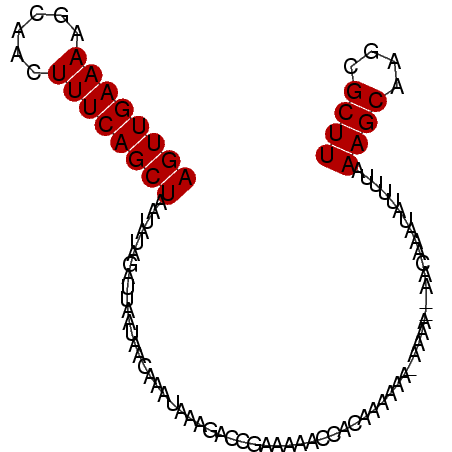

| Location | 8,115,938 – 8,116,038 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 83.81 |
| Mean single sequence MFE | -15.32 |
| Consensus MFE | -15.12 |
| Energy contribution | -15.15 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.53 |
| SVM RNA-class probability | 0.995022 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8115938 100 + 22407834 AAUAACA---------AAUAAAGACCGAAAAACCACAAAAA---AAA--AACAAAUAUUUUA-AAGCAAGCGCUUGAAUUGUACAUUUUUACAAGGCAACUGUACAUUAAGCGCC .......---------.........................---...--.............-......((((((((..((((((.........(....))))))))))))))). ( -15.60) >DroGri_CAF1 74665 108 + 1 AGUAAUUGUAACAUUUAAUAAAAACGGAAACA-----ACAA--AAAACCAAAAAAUAUUUUAAAAGCAAGCGCUUGAAUUGUACAUUUUUACAAGGCAACUGUACAUUUAGCGCC .........................(....).-----....--..........................(((((.((((.(((((.........(....))))))))))))))). ( -14.70) >DroSec_CAF1 71878 103 + 1 AAUAACA---------AAUAAAGACCGAAAAACCACAAAAAAAAAAA--AACAAAUAUUUUA-AAGCAAGCGCUUGAAUUGUACAUUUUUACAAGGCAACUGUACAUUAAGCGCC .......---------...............................--.............-......((((((((..((((((.........(....))))))))))))))). ( -15.60) >DroEre_CAF1 72559 94 + 1 AAUAACA---------AAUAAAGACCGAAAAACCACAAAAAA-----------AAUAUUUUA-AAGCAAGCGCUUGAAUUGUACAUUUUUACAAGGCAACUGUACAUUAAGCGCC .......---------..........................-----------.........-......((((((((..((((((.........(....))))))))))))))). ( -15.60) >DroYak_CAF1 73791 105 + 1 AAUAACA---------AAUAAAGACCGAAAAACCACAAAAAAAAAAAUCAACAAAUAUUUUA-AAGCAAGCGCUUGAAUUGUACAUUUUUACAAGGCAACUGUACAUUAAGCGCC .......---------..............................................-......((((((((..((((((.........(....))))))))))))))). ( -15.60) >DroAna_CAF1 74815 94 + 1 AAUAAUA---------AAAUCUGA--GAAAAAACCCAAAAAAA---------AAAUAUUUUA-AAGCAAGCGCUUGGAUUGUACAUUUUUACAAGGCAACUGUACAUUAAGCGCC .......---------........--.................---------..........-......((((((((..((((((.........(....))))))))))))))). ( -14.80) >consensus AAUAACA_________AAUAAAGACCGAAAAACCACAAAAAA__AAA__AACAAAUAUUUUA_AAGCAAGCGCUUGAAUUGUACAUUUUUACAAGGCAACUGUACAUUAAGCGCC .....................................................................((((((((..((((((.........(....))))))))))))))). (-15.12 = -15.15 + 0.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:37:24 2006