| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,113,708 – 8,113,814 |
| Length | 106 |
| Max. P | 0.755473 |

| Location | 8,113,708 – 8,113,814 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 80.36 |
| Mean single sequence MFE | -30.45 |
| Consensus MFE | -17.01 |
| Energy contribution | -16.33 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.755473 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
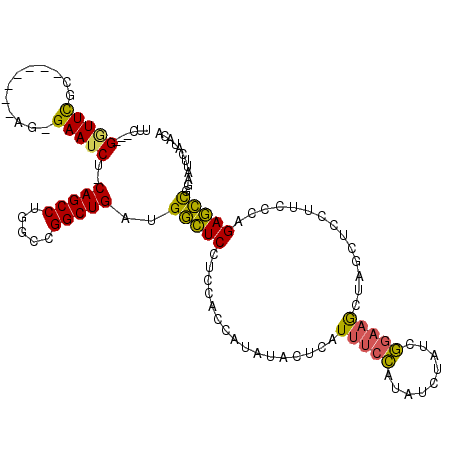
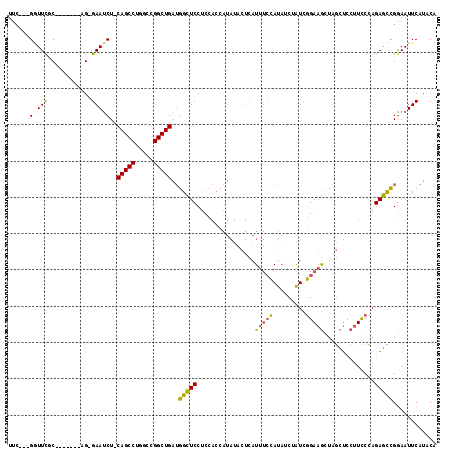
>2L_DroMel_CAF1 8113708 106 - 22407834 UUC---GGUUCGC-------AG-GAAUCU-CAGCCUGGCCGGCUGAUGGCUCCUCCACCAUAUACUCAUUUCCAUAUCUAUCGGAAGCUGGCUCCUUCCCAGAGCCGGAAUUCAUACA ...---(((..(.-------((-((..((-(((((.....)))))).)..))))).))).......................(((..(((((((.......)))))))..)))..... ( -33.30) >DroPse_CAF1 127998 96 - 1 UUG---GUUUUUCGGGUGGAAGAGAACCUGCAGCCUGGCUGGCUGGGGAUUCCCC------------AUU--AACAUCUGUCUGAAACUUA-----UCCUAGAAUUGGAAUUCCUACA ..(---(..(((((((..((..........(((((.....)))))(((....)))------------...--....))..)))))))....-----(((.......)))...)).... ( -26.90) >DroSec_CAF1 69646 106 - 1 UUC---GAUUCGC-------AG-GAAUCU-CAGCCUGGCCGGCUGAUGGCUCCUCCACCAUAUACUCAUUUCCAUAUCUAUCGGAAGCUAGCUCCUUCCCAGAGCCGGAAUUCAUACA ...---(((((((-------((-((..((-(((((.....)))))).)..))))...............((((.........))))))..((((.......))))..)))))...... ( -27.00) >DroSim_CAF1 69744 106 - 1 UUC---GGUUCGC-------AG-GAAUCU-CAGCCUGGCCGGCUGAUGGCUCCUCCACCAUAUACUCAUUUCCAUAUCUAUCGGAAGCUAGCUCCUUCCCAGAGCCGGAAUUCAUACA (((---((((((.-------((-((..((-(((((.....)))))).)..)))).)..........................(((((.......)))))..))))))))......... ( -33.10) >DroEre_CAF1 70333 105 - 1 UUC---GGUUCGC-------AG-GAAUCU-CAGCCUGGCCGGCUGAUGGCUCCUCCACCAUAUUCUCAUCUCCAUU-CGAUCGGAAGCUUGCUCCUUCCCAGAGCCGGAAUUCAUACA (((---((((((.-------((-((..((-(((((.....)))))).)..)))).)....................-.....(((((.......)))))..))))))))......... ( -33.10) >DroYak_CAF1 71541 109 - 1 UUCACAGGUUCGC-------AG-GAAUCU-CAGCCUGGCUGGCUGAUGGCUCCUCCACCAUAUACUCAUUUCCAUACCGAUCGGAAGCUAGCUCCUUUCCAGAGCCGAAAUUCAUACA ......((((((.-------((-((..((-(((((.....)))))).)..)))).)..........................(((((........))))).)))))............ ( -29.30) >consensus UUC___GGUUCGC_______AG_GAAUCU_CAGCCUGGCCGGCUGAUGGCUCCUCCACCAUAUACUCAUUUCCAUAUCUAUCGGAAGCUAGCUCCUUCCCAGAGCCGGAAUUCAUACA ......(((((............)))))..(((((.....)))))..(((((................(((((.........)))))..............)))))............ (-17.01 = -16.33 + -0.69)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:37:23 2006