
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,103,315 – 8,103,496 |
| Length | 181 |
| Max. P | 0.981078 |

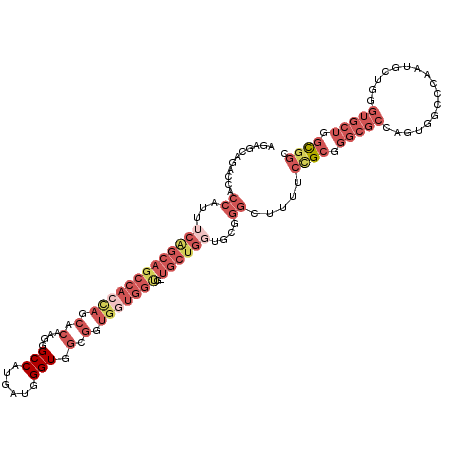
| Location | 8,103,315 – 8,103,433 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 67.85 |
| Mean single sequence MFE | -46.53 |
| Consensus MFE | -19.65 |
| Energy contribution | -23.91 |
| Covariance contribution | 4.27 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.42 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.521006 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8103315 118 + 22407834 AGAGCAGACCACCAUUUCAGCAGCCACCAGCACAAGGGCCAUGAUGGGUGGCGGUGGUGGCGUUGCUGGUGCGGGCUUUUCCGCGGGCGCCAGUGGCCCAAUGCUGGGUGCUGGCGGU ..........(((...((((((....((((((...(((((((....((((.(.((((.(((.((((....)))))))...)))).).)))).)))))))..)))))).)))))).))) ( -56.60) >DroSec_CAF1 59310 118 + 1 AGAGCAGACCACCAUUUCAGCAGCCACCAGCACAAGGGCCAUGAUGGGUGGCGGUGGUGGUGUUGCUGGUUCGGGCUUUUCCGCGGGCGCCAGUGGCCCUAUGCUGGGUGCUGGCGGU (((((......((...((((((((.((((.(((....(((((.....))))).))).))))))))))))...))))))).((((.((((((...(((.....))).)))))).)))). ( -54.00) >DroSim_CAF1 60748 118 + 1 AGAGCAGACCACCAUUUCAGCAGCCACUAGCACAAGGGCCAUGAUGGGUGGCGGUGGUGGUNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN .......(((((((((......((((((..((.........))...)))))))))))))))......................................................... ( -20.20) >DroEre_CAF1 59978 106 + 1 AGAGCAGACCACCAUUUCAGCAGCCACCAGCACAAGGGCCAUGAUGGGUGG------------UGCUGGUGCGGGCAUUUCUGCGGGCGCCAGUGGCCCAAUGCUGGGUGCUGGCGGC ...((.((........))..((((..((((((...(((((((......(((------------((((..((((((....))))))))))))))))))))..))))))..))))))... ( -46.80) >DroYak_CAF1 60940 118 + 1 AGAGCACACCACCAUUUCAGCAGCCACCAGCACAAGGGCCAUGAUGGGUGGCGGUGGUGGCGCUGCUGCUGCGGGCAUUUCUGCGGGCGCGAGUGGCCAAAUGCUGGGUGCUGGCGGC ..(((((.(((.((((((((((((..(((.(((....(((((.....))))).))).))).)))))))(((((((....)))))))((....))....))))).))))))))...... ( -51.20) >DroAna_CAF1 61695 100 + 1 GGAACAGACCACCAUAUCGGCAGCCACCAGCACAAUAGCCAUGUCUGGUCUGGGUGCUGGUGCUGCUGGUGGUGGCU---CCGU---------------GAUGAUGGGUGCUGUUAAC ..(((((.(((((((..(((((((.((((((((....((((....))))....)))))))))))))))))))))).(---((((---------------....)))))..)))))... ( -50.40) >consensus AGAGCAGACCACCAUUUCAGCAGCCACCAGCACAAGGGCCAUGAUGGGUGGCGGUGGUGGUG_UGCUGGUGCGGGCUUUUCCGCGGGCGCCAGUGGCCCAAUGCUGGGUGCUGGCGGC ...........((...(((((((((((((.(.(....(((......))).).).)))))))..))))))....)).....((((.(((((.................))))).)))). (-19.65 = -23.91 + 4.27)

| Location | 8,103,315 – 8,103,433 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 67.85 |
| Mean single sequence MFE | -42.38 |
| Consensus MFE | -17.01 |
| Energy contribution | -19.60 |
| Covariance contribution | 2.59 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.40 |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.981078 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8103315 118 - 22407834 ACCGCCAGCACCCAGCAUUGGGCCACUGGCGCCCGCGGAAAAGCCCGCACCAGCAACGCCACCACCGCCACCCAUCAUGGCCCUUGUGCUGGUGGCUGCUGAAAUGGUGGUCUGCUCU ((((((((..(((......)))...)))))(((.((((......))))..(((((..(((((((.(((...((.....)).....))).))))))))))))....))))))....... ( -48.70) >DroSec_CAF1 59310 118 - 1 ACCGCCAGCACCCAGCAUAGGGCCACUGGCGCCCGCGGAAAAGCCCGAACCAGCAACACCACCACCGCCACCCAUCAUGGCCCUUGUGCUGGUGGCUGCUGAAAUGGUGGUCUGCUCU ..((((((..(((......)))...))))))...(((((...(((.....(((((.(((((.(((.((((.......))))....))).)))))..)))))....)))..)))))... ( -47.50) >DroSim_CAF1 60748 118 - 1 NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNACCACCACCGCCACCCAUCAUGGCCCUUGUGCUAGUGGCUGCUGAAAUGGUGGUCUGCUCU ..................................................................((((((..((((((((((......)).))))).)))...))))))....... ( -17.90) >DroEre_CAF1 59978 106 - 1 GCCGCCAGCACCCAGCAUUGGGCCACUGGCGCCCGCAGAAAUGCCCGCACCAGCA------------CCACCCAUCAUGGCCCUUGUGCUGGUGGCUGCUGAAAUGGUGGUCUGCUCU (((((((((..(((((((.((((((((((.((..(((....)))..)).))))..------------..........))))))..)))))))..))........)))))))....... ( -47.50) >DroYak_CAF1 60940 118 - 1 GCCGCCAGCACCCAGCAUUUGGCCACUCGCGCCCGCAGAAAUGCCCGCAGCAGCAGCGCCACCACCGCCACCCAUCAUGGCCCUUGUGCUGGUGGCUGCUGAAAUGGUGGUGUGCUCU ......((((((((.(((((........(((...(((....))).)))..(((((((((((.(((.((((.......))))....))).)))).)))))))))))).))).))))).. ( -49.60) >DroAna_CAF1 61695 100 - 1 GUUAACAGCACCCAUCAUC---------------ACGG---AGCCACCACCAGCAGCACCAGCACCCAGACCAGACAUGGCUAUUGUGCUGGUGGCUGCCGAUAUGGUGGUCUGUUCC ...................---------------..((---(((.((((((((((((((((((((..((.(((....)))))...)))))))).))))).....)))))))..))))) ( -43.10) >consensus GCCGCCAGCACCCAGCAUUGGGCCACUGGCGCCCGCGGAAAAGCCCGCACCAGCA_CACCACCACCGCCACCCAUCAUGGCCCUUGUGCUGGUGGCUGCUGAAAUGGUGGUCUGCUCU ...((((((((........((((.......))))................................((((.......))))....))))))))(((..((.....))..)))...... (-17.01 = -19.60 + 2.59)

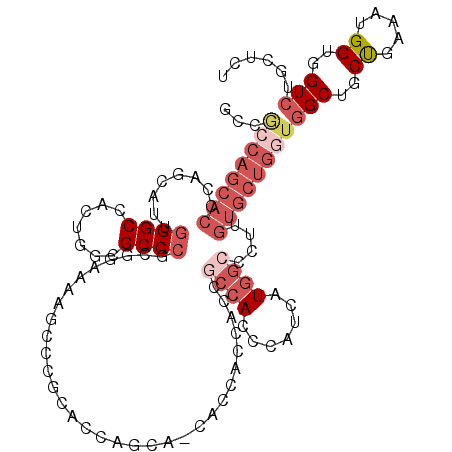

| Location | 8,103,393 – 8,103,496 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 81.26 |
| Mean single sequence MFE | -47.42 |
| Consensus MFE | -34.56 |
| Energy contribution | -38.72 |
| Covariance contribution | 4.16 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.889278 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8103393 103 + 22407834 UUCCGCGGGCGCCAGUGGCCCAAUGCUGGGUGCUGGCGGUCACAUGCUGAUGGGUGGCCAGGGGCAUCAGGUGCAUCUGCAGCACCAGACGCUCUUGCCCGUC ....(((((((((((((.((((....)))))))))))((((((..........))))))((((((.((.(((((.......))))).)).)))))))))))). ( -54.40) >DroSec_CAF1 59388 103 + 1 UUCCGCGGGCGCCAGUGGCCCUAUGCUGGGUGCUGGCGGUCACAUGCUGAUGGGUGGCCAGGGGCAUCAGGUGCAUCUGCAGCACCAGACGCUCUUGCCCGUC ....(((((((((((((.(((......))))))))))((((((..........))))))((((((.((.(((((.......))))).)).)))))))))))). ( -54.00) >DroEre_CAF1 60044 103 + 1 UUCUGCGGGCGCCAGUGGCCCAAUGCUGGGUGCUGGCGGCCACAUGCUGAUGGGUGGCCAGGGGCAUCAGGUGCAUCUGCAGCACCAGACGCUCUUGCCCGUC ....(((((((((((((.((((....)))))))))))((((((..........))))))((((((.((.(((((.......))))).)).)))))))))))). ( -57.00) >DroYak_CAF1 61018 103 + 1 UUCUGCGGGCGCGAGUGGCCAAAUGCUGGGUGCUGGCGGCCACAUGCUGAUGGGUGGCCAGGGGCAUCAGGUGCAUCUGCAGCACCAGACGCUCUUGCCCGUC ....((((((((.((((.(((.....))).)))).))((((((..........))))))((((((.((.(((((.......))))).)).)))))))))))). ( -51.60) >DroAna_CAF1 61772 80 + 1 --CCGU---------------GAUGAUGGGUGCUGUUAACCACAUGAUGAUGGGUGGU------GGUCAGUUGCUUCAGCCCUCCCAGACGCUCUUGCCUAUC --((((---------------.((.((((((.......))).))).)).))))..(((------((.(((..((.((..........)).))..))).))))) ( -20.10) >consensus UUCCGCGGGCGCCAGUGGCCCAAUGCUGGGUGCUGGCGGCCACAUGCUGAUGGGUGGCCAGGGGCAUCAGGUGCAUCUGCAGCACCAGACGCUCUUGCCCGUC ....(((((((((((((.(((......))))))))))((((((..........))))))((((((.((.(((((.......))))).)).)))))))))))). (-34.56 = -38.72 + 4.16)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:37:22 2006