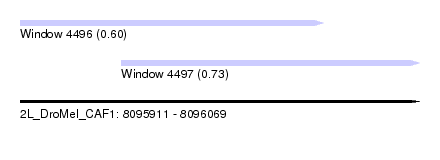
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,095,911 – 8,096,069 |
| Length | 158 |
| Max. P | 0.731002 |

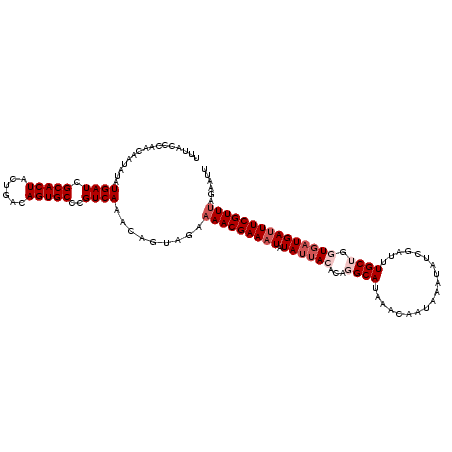
| Location | 8,095,911 – 8,096,031 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.73 |
| Mean single sequence MFE | -22.51 |
| Consensus MFE | -18.29 |
| Energy contribution | -19.89 |
| Covariance contribution | 1.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.602360 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8095911 120 + 22407834 UUUACCCAACAAUAUAUGAUCGCACUACUGACAGUGCCCGUCAAACAGUAGAAAACGAAAUAUAUUACACAGGCAUAAACAAUAAAUAUCGAUUUGCUGGUGAUGAUUUCGUUUAGAAUU ................((((.(((((......)))))..)))).........(((((((((.((((((...((((...................)))).)))))))))))))))...... ( -24.51) >DroSec_CAF1 52111 120 + 1 UUUACCCAACAAUAUAUGAUCGCACUACUGACAGUGCCCGUCAAACAGCAGAAAACGAAAUAUAUUACACAGGCAUAAACAAUAAAUAUCGAUUUGCUGGUGAUGAUUUCGUUUAGAAUU ................((((.(((((......)))))..)))).........(((((((((.((((((...((((...................)))).)))))))))))))))...... ( -24.51) >DroSim_CAF1 53336 120 + 1 UUUACCCAACAAUAUAUGAUCGCACUACUGACAGUGCCCGUCAAACAGUAGAAAACGAAAUAUAUUACACAGGCAUAAACAAUAAAUAUCGAUUUGCUGGUGAUGAAUUCGUUUAGAAUU ................((((.(((((......)))))..)))).........(((((((...((((((...((((...................)))).))))))..)))))))...... ( -21.31) >DroEre_CAF1 52629 115 + 1 UUUCCCCAACAA----UGAUCGCACUACUGUCAGUGC-CGUCAAACAGUAGAAAACGAAAUAUAUAACACAGGCAUAAACAAUAAAUAUCGAUUUGCAUAUGUUGAUUUCGUUUAGAAUU ............----((((.(((((......)))))-.)))).........(((((((((...(((((...(((...................)))...))))))))))))))...... ( -22.11) >DroYak_CAF1 53309 115 + 1 UUUGCCCAACAA----UGAUCGCACUACUGACAGUGCCCGUCAAACAGUAUA-AACGAAAUAUAUUACCCAGGCAUAAACAAUGAAUAUCGAUUUGCAGAUGAUGAUUUCGUUUAGAAUU ............----((((.(((((......)))))..)))).......((-((((((((.(((((.(...(((...................))).).)))))))))))))))..... ( -20.11) >consensus UUUACCCAACAAUAUAUGAUCGCACUACUGACAGUGCCCGUCAAACAGUAGAAAACGAAAUAUAUUACACAGGCAUAAACAAUAAAUAUCGAUUUGCUGGUGAUGAUUUCGUUUAGAAUU ................((((.(((((......)))))..)))).........(((((((((.((((((...((((...................)))).)))))))))))))))...... (-18.29 = -19.89 + 1.60)

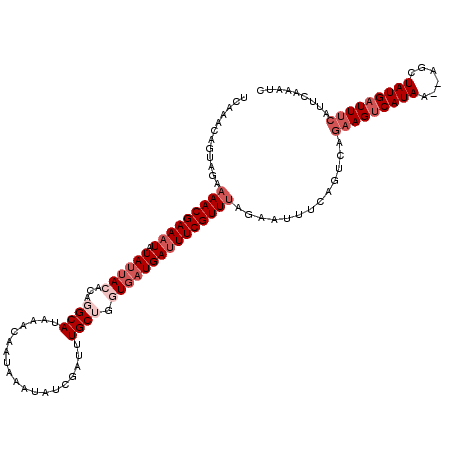
| Location | 8,095,951 – 8,096,069 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.09 |
| Mean single sequence MFE | -21.75 |
| Consensus MFE | -15.38 |
| Energy contribution | -17.93 |
| Covariance contribution | 2.55 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.731002 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8095951 118 + 22407834 UCAAACAGUAGAAAACGAAAUAUAUUACACAGGCAUAAACAAUAAAUAUCGAUUUGCUGGUGAUGAUUUCGUUUAGAAUUUCAGUCAGAAGUCAUAA--AGCUAUGAUUUCAUUCAAAGC ..........(((((((((((.((((((...((((...................)))).))))))))))))))).............(((((((((.--...)))))))))..))..... ( -25.81) >DroSec_CAF1 52151 116 + 1 UCAAACAGCAGAAAACGAAAUAUAUUACACAGGCAUAAACAAUAAAUAUCGAUUUGCUGGUGAUGAUUUCGUUUAGAAUUUCAGUCAGAAGUCAUAA--AGCUAUGAUUUCAUUUAUA-- ............(((((((((.((((((...((((...................)))).))))))))))))))).............(((((((((.--...))))))))).......-- ( -25.61) >DroSim_CAF1 53376 118 + 1 UCAAACAGUAGAAAACGAAAUAUAUUACACAGGCAUAAACAAUAAAUAUCGAUUUGCUGGUGAUGAAUUCGUUUAGAAUUUCAGUCAGAAGGCAUAA--AGCUAUGAUUUCAUUCAAAUC ............(((((((...((((((...((((...................)))).))))))..))))))).((((...(((((...(((....--.))).)))))..))))..... ( -20.91) >DroEre_CAF1 52664 120 + 1 UCAAACAGUAGAAAACGAAAUAUAUAACACAGGCAUAAACAAUAAAUAUCGAUUUGCAUAUGUUGAUUUCGUUUAGAAUUUCACGCAGAAGUCAUAUGUACAUAUGAUUUCAUACAAAUC .......(((..(((((((((...(((((...(((...................)))...)))))))))))))).............((((((((((....)))))))))).)))..... ( -24.21) >DroYak_CAF1 53345 97 + 1 UCAAACAGUAUA-AACGAAAUAUAUUACCCAGGCAUAAACAAUGAAUAUCGAUUUGCAGAUGAUGAUUUCGUUUAGAAUUUCAGACA----------------------UUAUUAAAAUC ..........((-((((((((.(((((.(...(((...................))).).)))))))))))))))............----------------------........... ( -12.21) >consensus UCAAACAGUAGAAAACGAAAUAUAUUACACAGGCAUAAACAAUAAAUAUCGAUUUGCUGGUGAUGAUUUCGUUUAGAAUUUCAGUCAGAAGUCAUAA__AGCUAUGAUUUCAUUCAAAUC ............(((((((((.((((((...((((...................)))).))))))))))))))).............(((((((((......)))))))))......... (-15.38 = -17.93 + 2.55)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:37:18 2006