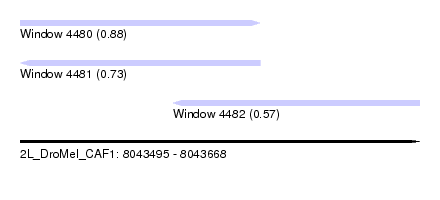
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,043,495 – 8,043,668 |
| Length | 173 |
| Max. P | 0.877861 |

| Location | 8,043,495 – 8,043,599 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 69.58 |
| Mean single sequence MFE | -28.30 |
| Consensus MFE | -15.37 |
| Energy contribution | -16.82 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.877861 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
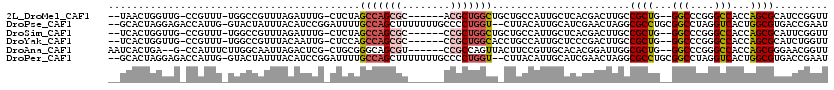
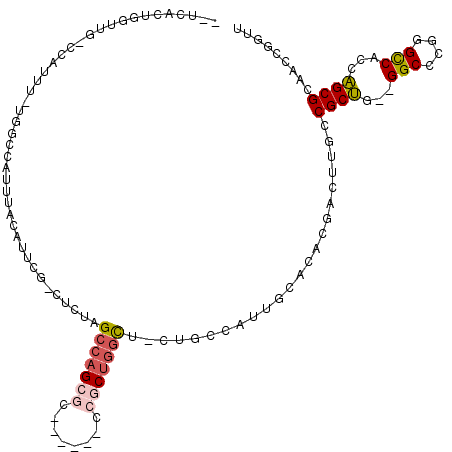
>2L_DroMel_CAF1 8043495 104 + 22407834 -------------AUCGAAAUAUUUUCAACAAUUUUCUGUGGAAAAUAAUUGGAAAUAUAGCGAAUCUC-CAGCAGCCGAAACCGGAUGCGCUGGUGGCCCGGGCC--CAGCGGCAAGUC -------------.(((..((((((((((..((((((....))))))..))))))))))..))).....-..(((.(((....))).)))((((.((((....).)--)).))))..... ( -35.80) >DroPse_CAF1 45752 119 + 1 CUGCUGUAAAUAAAUCGGAAUAUUUUCAAGAAGUUUUUAGCGAAAA-AAUUGCAAAUAAAACAAAUUUGUUCUCUGUCAAAUUCGGUCACGCCAGUGACCUAGGCCGCAGGCGCCUAGUU ..((((.((((......(((....))).....)))).))))((...-....((((((.......))))))......))......((((((....))))))((((((....).)))))... ( -25.72) >DroSim_CAF1 2 104 + 1 -------------AUCGAAAUAUUUUCAACAAUUUUCCGUGGAAAAUAAUUGGAAAUAUAGCGAAUAUC-CAGCAGCCCAAACCGAAUGCGCUGGUGGCCCGGGCC--CAGCGGCAAGUC -------------.(((..((((((((((..((((((....))))))..))))))))))..))).....-..(((..(......)..)))((((.((((....).)--)).))))..... ( -31.30) >DroYak_CAF1 502 109 + 1 --------AAAAAAUCGAAAUAUUUUCAACAAUUUUCUUGCGAAAAUAAUUGGAAAUAUAGCGAAUAUU-CAGCAGACAAAACCAGAUGCGCUGGUGGCCCGGGCC--CAGCGGCAAGUC --------......(((..((((((((((..((((((....))))))..))))))))))..))).....-..(((............)))((((.((((....).)--)).))))..... ( -29.30) >DroAna_CAF1 1803 117 + 1 CACCUGUAAAUAAAUCGGAAUAUUUUCAUCAAUUUUUUGCCAAAAAUUAUUGAAACCAAAGUGAUCCUC-AAGCUGUUGAAACCGUUCCCGCUGGUGGCCCGGGCC--CAGCGCCAAUCC ................(((((..((((((((((..(((.....)))..)))))......(((.......-..)))..)))))..)))))((((((..((....)))--)))))....... ( -23.90) >DroPer_CAF1 66464 119 + 1 CUGCUGUAAAUAAAUCGGAAUAUUUUCAACAAAUUUUUAGCGAAAA-AAUUGCAAAUAAAACAAAUUUGUUCUCUGUUAAAUUCGGUCACGCCAGUGACCUAGGCCGCAGGCGCCUAGUU .(((((.((((......(((....))).....)))).)))))....-....((((((.......))))))..............((((((....))))))((((((....).)))))... ( -23.80) >consensus ________AAUAAAUCGAAAUAUUUUCAACAAUUUUCUGGCGAAAAUAAUUGGAAAUAAAGCGAAUAUC_CAGCAGUCAAAACCGGACGCGCUGGUGGCCCGGGCC__CAGCGCCAAGUC ..............(((...(((((((((..((((((....))))))..)))))))))...))).........................(((((..(((....)))..)))))....... (-15.37 = -16.82 + 1.45)



| Location | 8,043,495 – 8,043,599 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 69.58 |
| Mean single sequence MFE | -30.08 |
| Consensus MFE | -16.51 |
| Energy contribution | -16.98 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.734222 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8043495 104 - 22407834 GACUUGCCGCUG--GGCCCGGGCCACCAGCGCAUCCGGUUUCGGCUGCUG-GAGAUUCGCUAUAUUUCCAAUUAUUUUCCACAGAAAAUUGUUGAAAAUAUUUCGAU------------- ....(((.((((--(((....))..))))))))((((((.......))))-))...(((..((((((.((((.((((((....)))))).)))).))))))..))).------------- ( -32.90) >DroPse_CAF1 45752 119 - 1 AACUAGGCGCCUGCGGCCUAGGUCACUGGCGUGACCGAAUUUGACAGAGAACAAAUUUGUUUUAUUUGCAAUU-UUUUCGCUAAAAACUUCUUGAAAAUAUUCCGAUUUAUUUACAGCAG ..........(((((((...((((((....))))))(((.(((.(((((((((....)))))..)))))))..-..))))))..........((.(((((........))))).)))))) ( -24.80) >DroSim_CAF1 2 104 - 1 GACUUGCCGCUG--GGCCCGGGCCACCAGCGCAUUCGGUUUGGGCUGCUG-GAUAUUCGCUAUAUUUCCAAUUAUUUUCCACGGAAAAUUGUUGAAAAUAUUUCGAU------------- ......((((..--((((((((((............)))))))))))).)-)....(((..((((((.((((.((((((....)))))).)))).))))))..))).------------- ( -32.80) >DroYak_CAF1 502 109 - 1 GACUUGCCGCUG--GGCCCGGGCCACCAGCGCAUCUGGUUUUGUCUGCUG-AAUAUUCGCUAUAUUUCCAAUUAUUUUCGCAAGAAAAUUGUUGAAAAUAUUUCGAUUUUUU-------- (((..(((((((--(((....))..)))))......)))...))).....-.....(((..((((((.((((.((((((....)))))).)))).))))))..)))......-------- ( -31.10) >DroAna_CAF1 1803 117 - 1 GGAUUGGCGCUG--GGCCCGGGCCACCAGCGGGAACGGUUUCAACAGCUU-GAGGAUCACUUUGGUUUCAAUAAUUUUUGGCAAAAAAUUGAUGAAAAUAUUCCGAUUUAUUUACAGGUG ...((((.((((--..((((.........))))..)))).))))..((((-(..(((....(((((((((.((((((((....)))))))).))))).....))))...)))..))))). ( -30.00) >DroPer_CAF1 66464 119 - 1 AACUAGGCGCCUGCGGCCUAGGUCACUGGCGUGACCGAAUUUAACAGAGAACAAAUUUGUUUUAUUUGCAAUU-UUUUCGCUAAAAAUUUGUUGAAAAUAUUCCGAUUUAUUUACAGCAG ..((((((.......))))))(((((....)))))(((((..((((((.......))))))..))))).((((-(((.....)))))))(((((.(((((........))))).))))). ( -28.90) >consensus GACUUGCCGCUG__GGCCCGGGCCACCAGCGCAACCGGUUUUGACAGCUG_GAAAUUCGCUAUAUUUCCAAUUAUUUUCGCCAAAAAAUUGUUGAAAAUAUUCCGAUUUAUU________ .......((((...(((....)))...)))).........................(((...(((((.((((.((((((....)))))).)))).)))))...))).............. (-16.51 = -16.98 + 0.48)


| Location | 8,043,561 – 8,043,668 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 66.69 |
| Mean single sequence MFE | -40.75 |
| Consensus MFE | -18.10 |
| Energy contribution | -18.35 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.44 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.571641 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8043561 107 - 22407834 --UAACUGGUUG-CCGUUU-UGGCCGUUUAGAUUUG-CUCUAGCCAGCGC------ACGCUGGCUGCUGCCAUUGCUCACGACUUGCCGCUG--GGCCCGGGCCACCAGCGCAUCCGGUU --.((((((.((-(.(((.-(((((..........(-(..((((((((..------..))))))))..))....(((((((......)).))--)))...)))))..)))))).)))))) ( -45.20) >DroPse_CAF1 45831 115 - 1 --GCACUAGGAGACCAUUG-GUACUAUUUACAUCCGGAUUUUGCCAGCUUUUUUUGCCCCUGGU--CUUACAUUGCAUCGAACUAGGCGCCUGCGGCCUAGGUCACUGGCGUGACCGAAU --(((....(((((((.((-(((..((((......))))..)))))((.......))...))))--)))....))).(((..((((((.......))))))(((((....)))))))).. ( -37.10) >DroSim_CAF1 68 107 - 1 --UCACUGGUUG-CCGUUU-UGGCCGUUUAGAUUUG-CUCUAGCCAGCGC------CCGCUGGCUGCUGCCAUUGCUCACGACUUGCCGCUG--GGCCCGGGCCACCAGCGCAUUCGGUU --..((((..((-(.(((.-(((((..........(-(..((((((((..------..))))))))..))....(((((((......)).))--)))...)))))..))))))..)))). ( -42.10) >DroYak_CAF1 573 107 - 1 --UCACUGGUUG-CCGUUU-UGGCCGUUUACAAUUG-CUCCAGCCAGCGC------CCGCUGGCACCUGCCAUUGCUCCCGACUUGCCGCUG--GGCCCGGGCCACCAGCGCAUCUGGUU --..((..(.((-(.....-.(((.(((..((((.(-(....((((((..------..))))))....)).)))).....)))..)))((((--(((....))..)))))))).)..)). ( -38.90) >DroAna_CAF1 1882 108 - 1 AAUCACUGA--G-CCAUUUCUUGGCAAUUAGACUCG-CUGCGGGCAGCGU------CCGCCAGUUACUUCCGUUGCACACGGAUUGGCGCUG--GGCCCGGGCCACCAGCGGGAACGGUU ....((((.--(-(((.....)))).......((((-(((..(((..((.------.(.(((((..(.(((((.....)))))..)..))))--))..)).)))..)))))))..)))). ( -44.10) >DroPer_CAF1 66543 115 - 1 --GCACUAGGAGACCAUUG-GUACUAUUUACAUCCGGAUUUUGCCAGCUUUUUUUGCCCCUGGU--CUUACAUUGCAUCGAACUAGGCGCCUGCGGCCUAGGUCACUGGCGUGACCGAAU --(((....(((((((.((-(((..((((......))))..)))))((.......))...))))--)))....))).(((..((((((.......))))))(((((....)))))))).. ( -37.10) >consensus __UCACUGGUUG_CCAUUU_UGGCCAUUUACAUUCG_CUCUAGCCAGCGC______CCGCUGGCU_CUGCCAUUGCACACGACUUGCCGCUG__GGCCCGGGCCACCAGCGCAACCGGUU ..........................................(((((((........))))))).......................((((...(((....)))...))))......... (-18.10 = -18.35 + 0.25)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:37:02 2006