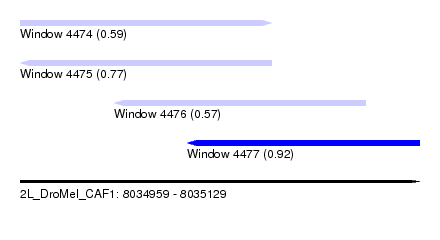
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,034,959 – 8,035,129 |
| Length | 170 |
| Max. P | 0.916504 |

| Location | 8,034,959 – 8,035,066 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 78.52 |
| Mean single sequence MFE | -24.22 |
| Consensus MFE | -16.80 |
| Energy contribution | -16.43 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.590236 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8034959 107 + 22407834 AAAUAAUAUGUAUAUCAAUAGCUAAGCACACAGCUAUUCAGCUGACCUGCAACGUUGGAGCGUGGGUGCUGACCACUUCCCCUCCAUUGUUGGUUUUAUGUUUUUUA (((.((((((...((((((((....(((..(((((....)))))...))).....(((((.(..((((.....))))..).)))))))))))))..)))))).))). ( -29.10) >DroSec_CAF1 2649 96 + 1 U---------CAUAUCAAUAACUAAGCA--CAGCUAUUCAACUGACCUGCAACGUUGGAGCGUGGGUGCUGACCACUUCCCCUCUAUUGUUGGUUUUAUGUUAUUUU .---------......((((((.((((.--(((........))).....(((((.(((((.(..((((.....))))..).))))).)))))))))...)))))).. ( -22.60) >DroSim_CAF1 2653 98 + 1 U---------CAUAUCAAUAACUAAGCACACAGCUAUUCAACUGACCUGCAACGUUGGAGCGUGGGUGCUGACCACUUCCCCUCCAUUGUUGGUUUUAUGUUAUUUU .---------......((((((.((((...(((........))).....(((((.(((((.(..((((.....))))..).))))).)))))))))...)))))).. ( -24.90) >DroYak_CAF1 2894 98 + 1 ACAUAAUACACAUAUCAAUCACUAAGCAGGCUGCUAUUUGACUGACCAGAAACGUAGAAGCGUGGGUGCUGCCCACUUCCCCUCCAUUUUUUGUGGU---------U ................((((((...((...((((..((((......))))...))))..))(((((.....)))))................)))))---------) ( -20.30) >consensus A_________CAUAUCAAUAACUAAGCACACAGCUAUUCAACUGACCUGCAACGUUGGAGCGUGGGUGCUGACCACUUCCCCUCCAUUGUUGGUUUUAUGUUAUUUU ...................(((..(((.....)))..............(((((.(((((.(..((((.....))))..).))))).))))))))............ (-16.80 = -16.43 + -0.37)

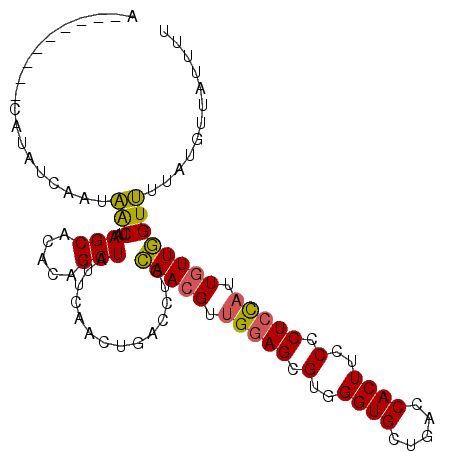
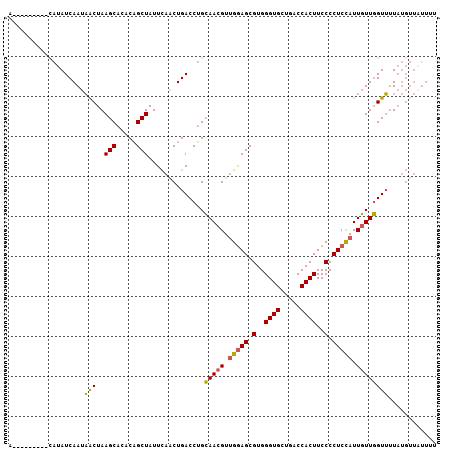
| Location | 8,034,959 – 8,035,066 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 78.52 |
| Mean single sequence MFE | -26.32 |
| Consensus MFE | -19.74 |
| Energy contribution | -19.30 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.769030 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8034959 107 - 22407834 UAAAAAACAUAAAACCAACAAUGGAGGGGAAGUGGUCAGCACCCACGCUCCAACGUUGCAGGUCAGCUGAAUAGCUGUGUGCUUAGCUAUUGAUAUACAUAUUAUUU .......((....(((..(((((..((((..((((.......)))).))))..)))))..)))....))((((((((......))))))))................ ( -26.60) >DroSec_CAF1 2649 96 - 1 AAAAUAACAUAAAACCAACAAUAGAGGGGAAGUGGUCAGCACCCACGCUCCAACGUUGCAGGUCAGUUGAAUAGCUG--UGCUUAGUUAUUGAUAUG---------A .......((((..(((..((((...((((..((((.......)))).))))...))))..)))....(.((((((((--....)))))))).)))))---------. ( -21.70) >DroSim_CAF1 2653 98 - 1 AAAAUAACAUAAAACCAACAAUGGAGGGGAAGUGGUCAGCACCCACGCUCCAACGUUGCAGGUCAGUUGAAUAGCUGUGUGCUUAGUUAUUGAUAUG---------A .......((((..(((..(((((..((((..((((.......)))).))))..)))))..)))....(.((((((((......)))))))).)))))---------. ( -26.30) >DroYak_CAF1 2894 98 - 1 A---------ACCACAAAAAAUGGAGGGGAAGUGGGCAGCACCCACGCUUCUACGUUUCUGGUCAGUCAAAUAGCAGCCUGCUUAGUGAUUGAUAUGUGUAUUAUGU .---------..((((..(((((.(((((..(((((.....))))).))))).)))))...((((((((...(((.....)))...)))))))).))))........ ( -30.70) >consensus AAAAUAACAUAAAACCAACAAUGGAGGGGAAGUGGUCAGCACCCACGCUCCAACGUUGCAGGUCAGUUGAAUAGCUGUGUGCUUAGUUAUUGAUAUG_________A ..................(((((..((((..((((.......)))).))))..)))))...((((((.(...(((.....)))...).))))))............. (-19.74 = -19.30 + -0.44)

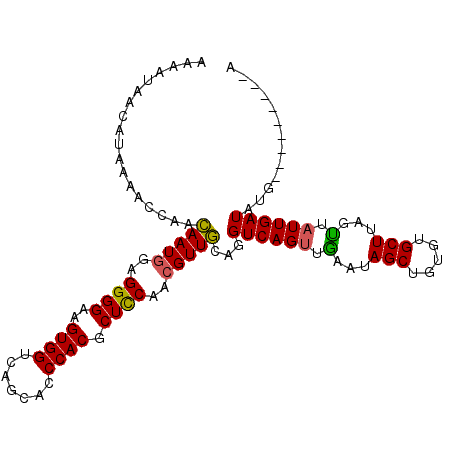
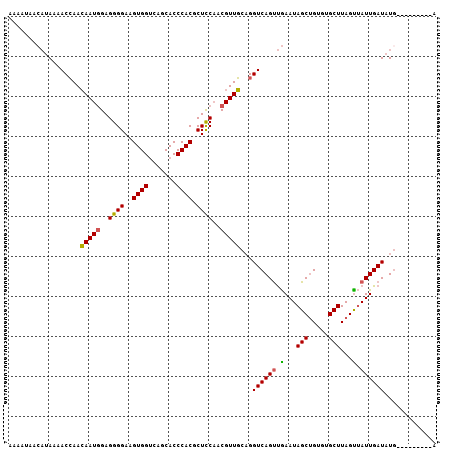
| Location | 8,034,999 – 8,035,106 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 85.51 |
| Mean single sequence MFE | -20.50 |
| Consensus MFE | -17.69 |
| Energy contribution | -17.38 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.570292 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8034999 107 - 22407834 CCAGAAGUAUAACUAUGACGCCCCUGCAAAAAAUAAAAAAUAAAAAACAUAAAACCAACAAUGGAGGGGAAGUGGUCAGCACCCACGCUCCAACGUUGCAGGUCAGC ...............(((....(((((((.........................(((....))).((((..((((.......)))).))))....)))))))))).. ( -22.30) >DroSec_CAF1 2678 107 - 1 CCAGAAGUAUAACUAUGACGCCCCAGCAAAAAAUAUAAAAAAAAUAACAUAAAACCAACAAUAGAGGGGAAGUGGUCAGCACCCACGCUCCAACGUUGCAGGUCAGU ...............((((.(..((((...........................((.........))(((.((((.......))))..)))...))))..))))).. ( -17.30) >DroSim_CAF1 2684 107 - 1 CCAGAAGUAUAACUAUGACGCCCCAGCAAAAAUAAUAAAAAAAAUAACAUAAAACCAACAAUGGAGGGGAAGUGGUCAGCACCCACGCUCCAACGUUGCAGGUCAGU ...............((((((....))...............................(((((..((((..((((.......)))).))))..)))))...)))).. ( -20.10) >DroYak_CAF1 2934 97 - 1 UCAGAACUAAAACUAUGACGCCCCUGCAAAAUAAA-AAAAA---------ACCACAAAAAAUGGAGGGGAAGUGGGCAGCACCCACGCUUCUACGUUUCUGGUCAGU ((((((..............(((((.((.......-.....---------...........)).)))))..(((((.....)))))..........))))))..... ( -22.30) >consensus CCAGAAGUAUAACUAUGACGCCCCAGCAAAAAAAAUAAAAAAAAUAACAUAAAACCAACAAUGGAGGGGAAGUGGUCAGCACCCACGCUCCAACGUUGCAGGUCAGU ...............((((((....))...............................(((((..((((..((((.......)))).))))..)))))...)))).. (-17.69 = -17.38 + -0.31)

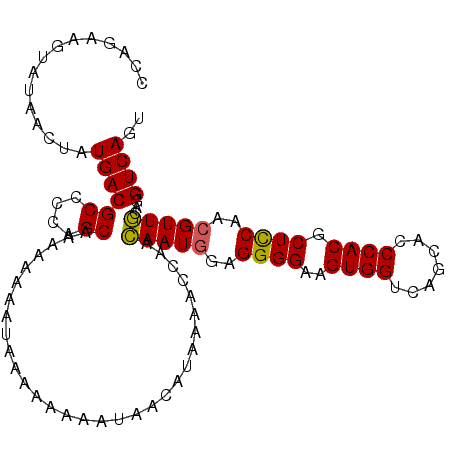
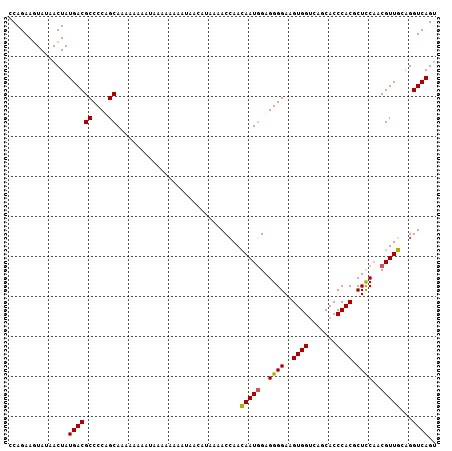
| Location | 8,035,030 – 8,035,129 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 85.52 |
| Mean single sequence MFE | -18.45 |
| Consensus MFE | -15.97 |
| Energy contribution | -16.54 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.916504 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8035030 99 - 22407834 UACUCUAACUGGCUAACUUUGUGCCAGAAGUAUAACUAUGACGCCCCUGCAAAAAAUAAAAAAUAAAAAACAUAAAACCAACAAUGGAGGGGAAGUGGU ((((....(((((.........))))).))))..(((((....(((((.((.................................)).)))))..))))) ( -20.81) >DroSec_CAF1 2709 99 - 1 UAUUCUAACUGGCUAACUUUUUGCCAGAAGUAUAACUAUGACGCCCCAGCAAAAAAUAUAAAAAAAAUAACAUAAAACCAACAAUAGAGGGGAAGUGGU ........(((((.........))))).......(((((....((((.........................................))))..))))) ( -16.48) >DroSim_CAF1 2715 99 - 1 UAUUCUAACUGGCUAACUUUUUGCCAGAAGUAUAACUAUGACGCCCCAGCAAAAAUAAUAAAAAAAAUAACAUAAAACCAACAAUGGAGGGGAAGUGGU ........(((((.........))))).......(((((....((((...........((.......))........(((....))).))))..))))) ( -18.30) >DroYak_CAF1 2965 89 - 1 UACUCUAACUGGCUAACUUUGUGUCAGAACUAAAACUAUGACGCCCCUGCAAAAUAAA-AAAAA---------ACCACAAAAAAUGGAGGGGAAGUGGG ........(((((.........)))))........((((....(((((.((.......-.....---------...........)).)))))..)))). ( -18.20) >consensus UACUCUAACUGGCUAACUUUGUGCCAGAAGUAUAACUAUGACGCCCCAGCAAAAAAAAUAAAAAAAAUAACAUAAAACCAACAAUGGAGGGGAAGUGGU ........(((((.........)))))........((((....(((((.((.................................)).)))))..)))). (-15.97 = -16.54 + 0.56)

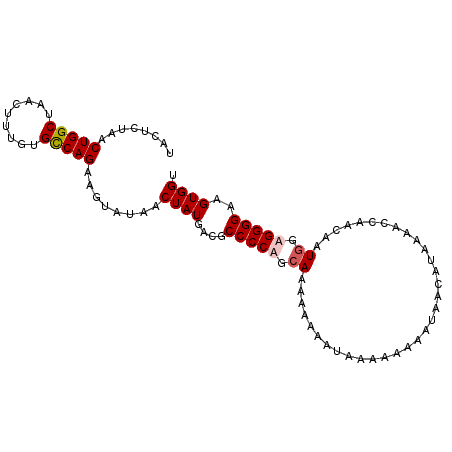
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:36:57 2006