| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,029,184 – 8,029,304 |
| Length | 120 |
| Max. P | 0.994722 |

| Location | 8,029,184 – 8,029,304 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.98 |
| Mean single sequence MFE | -21.86 |
| Consensus MFE | -10.92 |
| Energy contribution | -13.28 |
| Covariance contribution | 2.36 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.50 |
| SVM decision value | 2.50 |
| SVM RNA-class probability | 0.994722 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
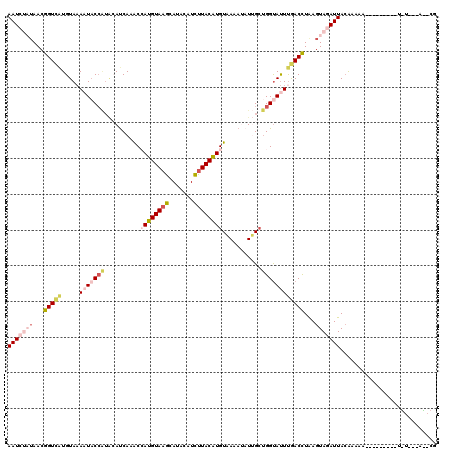
>2L_DroMel_CAF1 8029184 120 + 22407834 AAUCUAAAACGGUCAUGUAAAAUACCAUACAUCAAACCAUGUAAGCAUACAUCUUACAUGUAAAAUAUUGCUGGCAUUUGACCUAAGUAGAUUAAGUAGAUUACAAAAAUAUGUAAUCCG ((((((....(((((((((........))))......((((((((.......))))))))..................)))))....)))))).....(((((((......))))))).. ( -25.70) >DroSec_CAF1 6327 100 + 1 AAUCUUUAACGGUCAUGUACAAUUCCAUACAUCAAGCCAUGUAAGCAUUCAUUUUACAUGUAAAAUAUUGCUGGUACUUGACCUAAGUAGAUUACACAAA-------------------- (((((.....(((((.((((.......(((((......)))))((((...((((((....))))))..)))).)))).))))).....))))).......-------------------- ( -24.00) >DroSim_CAF1 6702 100 + 1 AAUCUUUAACGGUCAUGUAAACUACCAUACAUCAAGCCAUGUAAGCAUUCAUUUUACAUGUAAAAUAUUGCUGGUAUUUGACCUAAGUAGAUUACACAAA-------------------- (((((.....(((((.......((((.(((((......)))))((((...((((((....))))))..))))))))..))))).....))))).......-------------------- ( -20.60) >DroEre_CAF1 4832 106 + 1 AAUCCAUAACGGUUGUGUAAAAUAC-----AUCAAACAAUGUAAGUAUACUUCCUACGUGUAAUAGUUCGGUGGUAUUUAACUUGAGUAAAUUACAAAAA---------UGUGUGAUGCG .(((((((..((..(((((...(((-----((......)))))..)))))..))....((((((..((((((........)).))))...))))))....---------)))).)))... ( -18.10) >DroYak_CAF1 7078 111 + 1 AAUCCAUAACGGUCAUGUAAAAUACCGUGGACCAAACAAUGUAAGUAUACAUCUUACAUGUCAUAAUUGGCCGGUAUUCAACUUAAGUAAAUUACAAAAA---------UGUCCUACGCG ..........((.(((((..((((((..((.(((((((.((((((.......))))))))).....))))))))))))..))....(((...)))....)---------)).))...... ( -20.90) >consensus AAUCUAUAACGGUCAUGUAAAAUACCAUACAUCAAACCAUGUAAGCAUACAUCUUACAUGUAAAAUAUUGCUGGUAUUUGACCUAAGUAGAUUACAAAAA_________U_U___A__CG (((((((...(((((.....(((((((..........((((((((.......))))))))...........))))))))))))...)))))))........................... (-10.92 = -13.28 + 2.36)


| Location | 8,029,184 – 8,029,304 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.98 |
| Mean single sequence MFE | -24.16 |
| Consensus MFE | -14.76 |
| Energy contribution | -15.92 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.970213 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
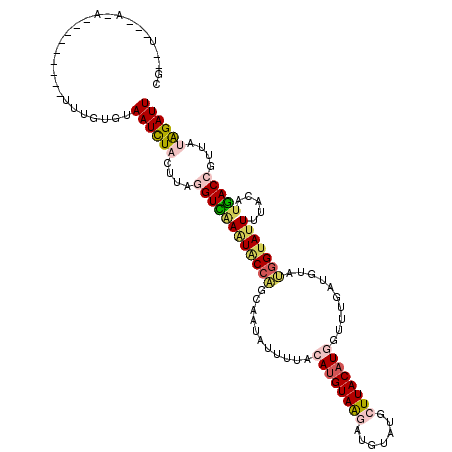
>2L_DroMel_CAF1 8029184 120 - 22407834 CGGAUUACAUAUUUUUGUAAUCUACUUAAUCUACUUAGGUCAAAUGCCAGCAAUAUUUUACAUGUAAGAUGUAUGCUUACAUGGUUUGAUGUAUGGUAUUUUACAUGACCGUUUUAGAUU .((((((((......))))))))....((((((....((((((((((((...(((((...((((((((.......))))))))....))))).))))))).....)))))....)))))) ( -37.20) >DroSec_CAF1 6327 100 - 1 --------------------UUUGUGUAAUCUACUUAGGUCAAGUACCAGCAAUAUUUUACAUGUAAAAUGAAUGCUUACAUGGCUUGAUGUAUGGAAUUGUACAUGACCGUUAAAGAUU --------------------.......(((((.....(((((.((((.((((.(((((((....)))))))..))))(((((......))))).......)))).))))).....))))) ( -22.80) >DroSim_CAF1 6702 100 - 1 --------------------UUUGUGUAAUCUACUUAGGUCAAAUACCAGCAAUAUUUUACAUGUAAAAUGAAUGCUUACAUGGCUUGAUGUAUGGUAGUUUACAUGACCGUUAAAGAUU --------------------.(..(((((.(((((((.(((((...((((((.(((((((....)))))))..))).....))).))))).)).))))).)))))..)............ ( -22.30) >DroEre_CAF1 4832 106 - 1 CGCAUCACACA---------UUUUUGUAAUUUACUCAAGUUAAAUACCACCGAACUAUUACACGUAGGAAGUAUACUUACAUUGUUUGAU-----GUAUUUUACACAACCGUUAUGGAUU .((((((.(((---------....(((((..(((((..((.....)).......((((.....))))).))))...))))).))).))))-----))...........((.....))... ( -14.70) >DroYak_CAF1 7078 111 - 1 CGCGUAGGACA---------UUUUUGUAAUUUACUUAAGUUGAAUACCGGCCAAUUAUGACAUGUAAGAUGUAUACUUACAUUGUUUGGUCCACGGUAUUUUACAUGACCGUUAUGGAUU ..(((((((((---------....)))...........((.((((((((((((.....((((((((((.......)))))).))))))))...)))))))).))....))..)))).... ( -23.80) >consensus CG__U___A_A_________UUUGUGUAAUCUACUUAGGUCAAAUACCAGCAAUAUUUUACAUGUAAGAUGUAUGCUUACAUGGUUUGAUGUAUGGUAUUUUACAUGACCGUUAUAGAUU ...........................((((((....((((((((((((...........((((((((.......))))))))..........))))))).....)))))....)))))) (-14.76 = -15.92 + 1.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:36:54 2006