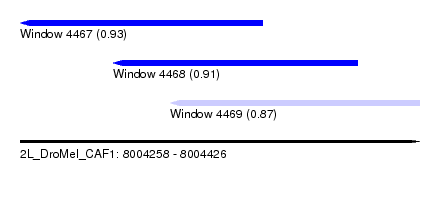
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,004,258 – 8,004,426 |
| Length | 168 |
| Max. P | 0.925497 |

| Location | 8,004,258 – 8,004,360 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.24 |
| Mean single sequence MFE | -29.54 |
| Consensus MFE | -21.11 |
| Energy contribution | -21.99 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.925497 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8004258 102 - 22407834 GCUGAU-UAUUAGCGGGUAACAUGUUUCGUUUAACGGAACAGCUGUUCAU-UUAG---------------UCGGCAGUUAUCGCUUAAGCCGCAUGUUAUCGAUAUCUUCAAU-UAUUUU (((((.-..))))).((((((((((...((((((((((((.((((.....-....---------------.)))).))).))).)))))).))))))))))............-...... ( -26.60) >DroSec_CAF1 4880 102 - 1 GCUGUU-UAUUAACGGGUAACAUGCUUCGUUUAACGGCACAGCUGUUCAU-UUAG---------------UCGGCAGUUAUCGGUUAAGCCGCAUGUUAUCGAUAUCUUCAAU-UAUUUU .(((((-....)))))(((((((((...(((((((.(...(((((((...-....---------------..)))))))..).))))))).))))))))).............-...... ( -25.90) >DroSim_CAF1 4829 102 - 1 GCUGUU-UAUUAACGGGUAACAUGCUUCGUUUAACGGAACAGCUGUUCAU-UUAG---------------ACGGCAGUUAUCGGUUAAGCCGCAUGCUAUCGAUAUCUUCAAU-UAUUUU (((((.-..(((((.((((((.(((..(((((((..((((....))))..-))))---------------)))))))))))).)))))...))).))................-...... ( -28.90) >DroEre_CAF1 4742 104 - 1 GCUGUUCUUUUAGCGGGUAACAUGCAUCGCUUAACGGAACAGCUGUUACU-UCAG---------------UCGGCCGUUAUCGGUUAAGCCGCAUGUUAUCGAUACCCGGAAUUUAUUUU ((((......)))).((((((((((...(((((((.((((.((((.....-....---------------.)))).))..)).))))))).))))))))))................... ( -33.40) >DroYak_CAF1 4928 120 - 1 GCUUUUUUUUUAACGGGUAACAUGCAUCGUUUAACGGAACAGCUGUUAUUUUCAGUUACAGAACUGCUGUUCGGCUGUUAUGGGCAAAGCCGCAUGUUAUCGAUAACCGAAAUUUAUUUG ...............((((((((((.........((.((((((((..((...(((((....)))))..)).)))))))).))(((...)))))))))))))................... ( -32.90) >consensus GCUGUU_UAUUAACGGGUAACAUGCUUCGUUUAACGGAACAGCUGUUCAU_UUAG_______________UCGGCAGUUAUCGGUUAAGCCGCAUGUUAUCGAUAUCUUCAAU_UAUUUU ...............((((((((((...(((((((.((((.((((..........................)))).))..)).))))))).))))))))))................... (-21.11 = -21.99 + 0.88)


| Location | 8,004,297 – 8,004,400 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.74 |
| Mean single sequence MFE | -32.70 |
| Consensus MFE | -19.71 |
| Energy contribution | -19.99 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.909808 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8004297 103 - 22407834 AAUCAACCGGUUGCGGUUAAAAGCUCUGUUAAACGGAGAUGCUGAU-UAUUAGCGGGUAACAUGUUUCGUUUAACGGAACAGCUGUUCAU-UUAG---------------UCGGCAGUUA ....(((((....))))).....(((((.....))))).(((((((-((..(((((......(((((((.....))))))).)))))...-.)))---------------)))))).... ( -31.30) >DroSec_CAF1 4919 103 - 1 AAUCAACCGGUUGCGAUUAAAAGCUCUGUUAAAUUGAGAAGCUGUU-UAUUAACGGGUAACAUGCUUCGUUUAACGGCACAGCUGUUCAU-UUAG---------------UCGGCAGUUA ..........((((((((((((((((((((((((...(((((((((-.(((....))))))).))))))))))))))...)))).....)-))))---------------)).))))... ( -25.50) >DroSim_CAF1 4868 103 - 1 AAUCAACCGGGUGCGGUUAAAAGCUCUGUUAAAUGGAGAAGCUGUU-UAUUAACGGGUAACAUGCUUCGUUUAACGGAACAGCUGUUCAU-UUAG---------------ACGGCAGUUA ....(((((....)))))...((((((((((((((((..(((((((-(...(((((((.....)).))))).....)))))))).)))))-)).)---------------)))).)))). ( -33.00) >DroEre_CAF1 4782 103 - 1 AGCCAACCGGUUGCGGUUAACUGCUCUGUUAGGU-AAGACGCUGUUCUUUUAGCGGGUAACAUGCAUCGCUUAACGGAACAGCUGUUACU-UCAG---------------UCGGCCGUUA ((((....))))(((((..((((.......((((-((.(.((((((((.(((((((((.....)).)))).))).))))))))).)))))-))))---------------)..))))).. ( -37.61) >DroYak_CAF1 4968 120 - 1 AAACAACCGGUUGCGGUUAACUGCGCUGUUAAGCGUAGAUGCUUUUUUUUUAACGGGUAACAUGCAUCGUUUAACGGAACAGCUGUUAUUUUCAGUUACAGAACUGCUGUUCGGCUGUUA .((((.((((..((((((..(((..(((((((((...(((((.....................))))))))))))))..)))((((.((.....)).))))))))))...)))).)))). ( -36.10) >consensus AAUCAACCGGUUGCGGUUAAAAGCUCUGUUAAAUGGAGAAGCUGUU_UAUUAACGGGUAACAUGCUUCGUUUAACGGAACAGCUGUUCAU_UUAG_______________UCGGCAGUUA ....(((((....)))))....(.((((((((((...(((((((((............)))).))))))))))))))).).((((..........................))))..... (-19.71 = -19.99 + 0.28)


| Location | 8,004,321 – 8,004,426 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.00 |
| Mean single sequence MFE | -27.64 |
| Consensus MFE | -21.40 |
| Energy contribution | -21.44 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.867756 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8004321 105 - 22407834 -------------UCAUUUAAAUUUAAA-AAACAACCGCCAAUCAACCGGUUGCGGUUAAAAGCUCUGUUAAACGGAGAUGCUGAU-UAUUAGCGGGUAACAUGUUUCGUUUAACGGAAC -------------...............-....((((((.((((....))))))))))......((((((((((((((.((((((.-..)))))).........)))))))))))))).. ( -29.50) >DroSec_CAF1 4943 105 - 1 -------------UCAUUGAAAUUUUAA-AAACAACCGCCAAUCAACCGGUUGCGAUUAAAAGCUCUGUUAAAUUGAGAAGCUGUU-UAUUAACGGGUAACAUGCUUCGUUUAACGGCAC -------------.........((((((-...((((((.........))))))...))))))(((..(((((((...(((((((((-.(((....))))))).))))))))))))))).. ( -22.70) >DroSim_CAF1 4892 105 - 1 -------------UCAUUUAAAUUUUAA-AAACAACCGCCAAUCAACCGGGUGCGGUUAAAAGCUCUGUUAAAUGGAGAAGCUGUU-UAUUAACGGGUAACAUGCUUCGUUUAACGGAAC -------------...............-....((((((...((....))..))))))......((((((((((((((.(.(((((-....))))).)......)))))))))))))).. ( -26.90) >DroEre_CAF1 4806 119 - 1 CUGCCGGUACACCACAUUUAAAUUUAAAUAAACAACCGCCAGCCAACCGGUUGCGGUUAACUGCUCUGUUAGGU-AAGACGCUGUUCUUUUAGCGGGUAACAUGCAUCGCUUAACGGAAC ..((.((....))....................((((((.((((....))))))))))....))((((((((((-....(((((......))))).((.....))...)))))))))).. ( -33.00) >DroYak_CAF1 5008 120 - 1 UUAUUAGUACACCACAUUUAAAUUGAAAUAAAGAACCGCCAAACAACCGGUUGCGGUUAACUGCGCUGUUAAGCGUAGAUGCUUUUUUUUUAACGGGUAACAUGCAUCGUUUAACGGAAC ...........((...........((((.((((((((((..(((.....)))))))))..(((((((....)))))))...)))).)))).(((((((.....)).)))))....))... ( -26.10) >consensus _____________UCAUUUAAAUUUAAA_AAACAACCGCCAAUCAACCGGUUGCGGUUAAAAGCUCUGUUAAAUGGAGAAGCUGUU_UAUUAACGGGUAACAUGCUUCGUUUAACGGAAC .................................((((((.((((....)))))))))).......(((((((((...(((((((((............)))).))))))))))))))... (-21.40 = -21.44 + 0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:36:50 2006