
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 7,999,832 – 7,999,945 |
| Length | 113 |
| Max. P | 0.726446 |

| Location | 7,999,832 – 7,999,945 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 80.38 |
| Mean single sequence MFE | -35.53 |
| Consensus MFE | -25.39 |
| Energy contribution | -25.92 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.726446 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
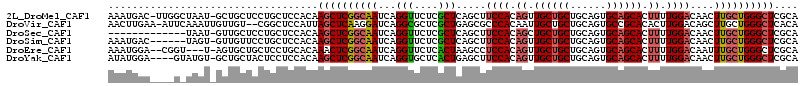
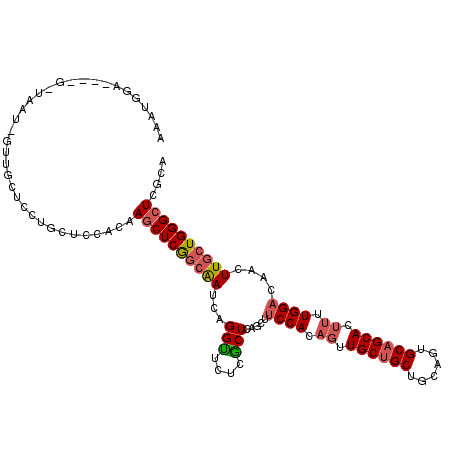
>2L_DroMel_CAF1 7999832 113 + 22407834 AAAUGAC-UUGGCUAAU-GCUGCUCCUGCUCCACAAGCUCGGCAAUCAGGUUCUCGCUCAGCUUCCACAGUUGCUGCUGCAGUGCAGCACUUUUGGACAACUUGCUGGGCUCGCA ....(((-(((((....-))((((...(((.....)))..))))..))))))...(((((((.((((.((.((((((......)))))).)).))))......)))))))..... ( -37.80) >DroVir_CAF1 463 112 + 1 AACUUGAA-AUUCAAAUUGUUGU--CGGCUCCAUUAGCUCAAGGAUCAGGCGCUCGCUGAGCGCCCACAAUUGCUGCUGCAGUGCCGCACACUUGGACAGCUUGCUGGGCUCACA ........-...........(((--.(((.(((..((((((((.....(((((((...)))))))......(((.((......)).)))..))))...))))...)))))).))) ( -33.50) >DroSec_CAF1 560 101 + 1 -------------UAAU-GUUGCUCCUGCUCCACAAGCUCGGCAAUCAGGUUCUCGCUCAGCUUCCACAGCUGCUGCUGCAGUGCAGCACUUUUGGACAACUUGCUGGGCUCGCA -------------....-((((((...(((.....)))..)))))).........(((((((.((((.((.((((((......)))))).)).))))......)))))))..... ( -33.60) >DroSim_CAF1 524 108 + 1 AAAUGAC------UAGU-GUUGUUCCUGCUCCACAAGCUCGGCAAUCAGGUUCUCGCUCAGCUUCCACAGUUGCUGCUGCAGUGCAGCACUUUUGGACAACUUGCUGGGCUCGCA .......------..((-(..(..((((..((........))....))))..)..(((((((.((((.((.((((((......)))))).)).))))......))))))).))). ( -33.40) >DroEre_CAF1 598 109 + 1 AAAUGGA--CGGU---U-AGUGCUGCUCCUGCACAAACUCGGCAAUCAGGUUCUCACUAAGCCUCCACAGUUGCUGCUGCAGUGCAGCACUUUUGGACAAUUUGCUGGGCUCGCA ....(((--((((---.-...)))).))).((.....((((((((...((((.......))))((((.((.((((((......)))))).)).))))....))))))))...)). ( -34.40) >DroYak_CAF1 625 110 + 1 AUAUGGA----GUAUGU-GCUGCUACUCCUCCACAAGCUCGGCAAUCAGGUGCUCACUGAGCUUCCACAGUUGCUGCUGCAGUGCAGCACUUUUGGACAACUUGCUGGGCUCGCA ....(((----(((.(.-....)))))))......((((((((((((((.......))))...((((.((.((((((......)))))).)).))))....)))))))))).... ( -40.50) >consensus AAAUGGA____G_UAAU_GUUGCUCCUGCUCCACAAGCUCGGCAAUCAGGUUCUCGCUCAGCUUCCACAGUUGCUGCUGCAGUGCAGCACUUUUGGACAACUUGCUGGGCUCGCA ...................................((((((((((...(((....))).....((((.((.((((((......)))))).)).))))....)))))))))).... (-25.39 = -25.92 + 0.53)
| Location | 7,999,832 – 7,999,945 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 80.38 |
| Mean single sequence MFE | -36.22 |
| Consensus MFE | -26.00 |
| Energy contribution | -26.12 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.558816 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
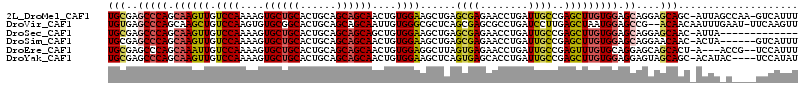
>2L_DroMel_CAF1 7999832 113 - 22407834 UGCGAGCCCAGCAAGUUGUCCAAAAGUGCUGCACUGCAGCAGCAACUGUGGAAGCUGAGCGAGAACCUGAUUGCCGAGCUUGUGGAGCAGGAGCAGC-AUUAGCCAA-GUCAUUU (((..((.((((......((((....((((((......))))))....)))).)))).)).....((((....(((......)))..)))).)))((-....))...-....... ( -35.10) >DroVir_CAF1 463 112 - 1 UGUGAGCCCAGCAAGCUGUCCAAGUGUGCGGCACUGCAGCAGCAAUUGUGGGCGCUCAGCGAGCGCCUGAUCCUUGAGCUAAUGGAGCCG--ACAACAAUUUGAAU-UUCAAGUU (((..(((((((..(((((...((((.....))))...)))))......((((((((...)))))))).........))...))).))..--)))..((((((...-..)))))) ( -37.30) >DroSec_CAF1 560 101 - 1 UGCGAGCCCAGCAAGUUGUCCAAAAGUGCUGCACUGCAGCAGCAGCUGUGGAAGCUGAGCGAGAACCUGAUUGCCGAGCUUGUGGAGCAGGAGCAAC-AUUA------------- (((.......))).(((((((.....((((((......))))))(((.((.(((((..((((........))))..))))).)).))).))).))))-....------------- ( -37.00) >DroSim_CAF1 524 108 - 1 UGCGAGCCCAGCAAGUUGUCCAAAAGUGCUGCACUGCAGCAGCAACUGUGGAAGCUGAGCGAGAACCUGAUUGCCGAGCUUGUGGAGCAGGAACAAC-ACUA------GUCAUUU (((.......))).(((((((.....((((((......)))))).((.((.(((((..((((........))))..))))).)).))..)).)))))-....------....... ( -32.60) >DroEre_CAF1 598 109 - 1 UGCGAGCCCAGCAAAUUGUCCAAAAGUGCUGCACUGCAGCAGCAACUGUGGAGGCUUAGUGAGAACCUGAUUGCCGAGUUUGUGCAGGAGCAGCACU-A---ACCG--UCCAUUU (((.......)))...........((((((((..(((....))).(((((.(((((..(..(........)..)..))))).)))))..))))))))-.---....--....... ( -33.70) >DroYak_CAF1 625 110 - 1 UGCGAGCCCAGCAAGUUGUCCAAAAGUGCUGCACUGCAGCAGCAACUGUGGAAGCUCAGUGAGCACCUGAUUGCCGAGCUUGUGGAGGAGUAGCAGC-ACAUAC----UCCAUAU .((((((.(.((((....((((....((((((......))))))....)))).((((...))))......)))).).))))))...((((((.....-...)))----))).... ( -41.60) >consensus UGCGAGCCCAGCAAGUUGUCCAAAAGUGCUGCACUGCAGCAGCAACUGUGGAAGCUGAGCGAGAACCUGAUUGCCGAGCUUGUGGAGCAGGAGCAAC_AUUA_C____UCCAUUU (((..(((((.((((((.((((....((((((......))))))....))))......((((........))))..))))))))).))....))).................... (-26.00 = -26.12 + 0.11)
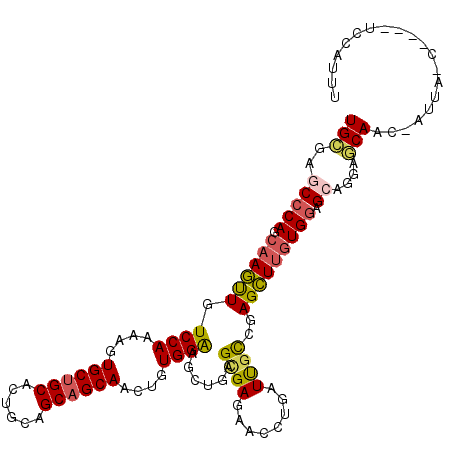
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:36:47 2006